AI scientist develops platform to track changes in genetic structure of COVID-19 virus

The COVID-19 virus is constantly changing as it moves from person to person. To fight the virus today and prepare for what it may look like tomorrow, researchers need to be able to track its development.
A team of healthcare professionals, led by Dr. Bo Wang, Artificial Intelligence (AI) Lead at the Peter Munk Cardiac Centre, have designed an innovative tool which enables researchers to track changes in the genetic structure of the SARS-CoV-2 virus, responsible for the onset of the COVID-19 global pandemic.
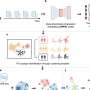
Developed by Dr. Wang's Ph.D. student, Hassaan Maan, alongside colleagues from McMaster University and Sunnybrook Health Sciences Centre, the COVID-19 Genotyping Tool (CGT) offers an online, user-friendly platform where researchers can compare the genome sequence of the SARS-CoV-2 virus in their hospital against the global picture.
Before development of the CGT, analysis of the virus could take up to a week. Now, using the AI-driven platform, this analysis takes minutes.
By following these changes, researchers can learn more about how the virus is moving and evolving, which has direct implications for vaccine design, drug development, and our collective effort to combat COVID-19.
Using nasal swab samples from more than 20,000 patients with COVID-19, virus genome sequences were uploaded to the Global Initiative on Sharing All Influenza Data (GISAID) COVID-19 database. The CGT then compares the sequence of the virus in individual hospitals with virus samples obtained from around the world. Results provide researchers with insight on where transmission events likely occurred, when outbreaks happened, and most importantly, alert them of any key changes in the genetic makeup of the virus, which determines how infectious it is.
"As AI researchers, we're accustomed to working with big data," says Dr. Wang, who also holds a Canadian Institute for Advanced Research in Artificial Intelligence Chair at the Vector Institute, and is an Assistant Professor at the University of Toronto Faculty of Medicine.
"The global research community has really embraced the initiative on data sharing during the COVID-19 pandemic; our goal was to design a tool that would help make sense of it."
The CGT is particularly helpful when preparing for a second wave of virus spread, which is often more deadly than the first. By keeping track of mutations in the SARS-CoV-2 virus, the CGT is helping researchers prepare to handle changes that can affect the severity of disease, impact of vaccines, and transmissibility of the virus.
"As the virus spreads throughout the world, it picks up small mutations along the way," says Hassaan Maan, a Ph.D. student in Dr. Wang's lab. "We want to perform surveillance of these mutations in the event a different strain of SARS-CoV-2 arises."
The speed of analysis and volume of data are defining characteristics of the application. The platform quickly processes user-uploaded data and offers immediate insights into the genomic evolution, generating results in less than 15 minutes.
The tool is purposely designed to be open source, as worldwide access will help researchers and healthcare professionals usher an end to this pandemic as quickly as possible. Dr. Wang and his team are funded thanks to the generous support of donors.
More information: Hassaan Maan et al, Genotyping SARS-CoV-2 through an interactive web application, The Lancet Digital Health (2020). DOI: 10.1016/S2589-7500(20)30140-0