Subtypes and developmental pathways of innate T cells identified
There are memory cells that remember previously encountered pathogens and help to react quickly and strongly when exposed to them again. The developmental process of strong immune cells that make these memory cells in advance without having to encounter the pathogens have been discovered.
Understanding the developmental process of these cells, which are responsible for biodefense in places where contact with pathogens is common such as the lungs, intestines and skin is anticipated to be used as fundamental data to overcome various infectious diseases or malignant tumors caused by immune dysregulation.
The National Research Foundation of Korea has announced that the joint research team from the Korea Advanced Institute of Science and Technology (KAIST) and Yonsei University Health Systems—led by professors You Jeong Lee and Sanguk Kim of Pohang Institute of Science and Technology (POSTECH) and Professor Jong Kyoung Kim of Daegu Gyeongbuk Institute of Science and Technology (DGIST)—have identified the developmental process of novel immune T cells. These research findings were published in the international journal Nature Communications on August 31.
T cells, which play an essential role in eliminating pathogens and cancer cells such as viruses, germs and fungi, including the recently prevalent COVID-19, have more than 10 different subtypes.
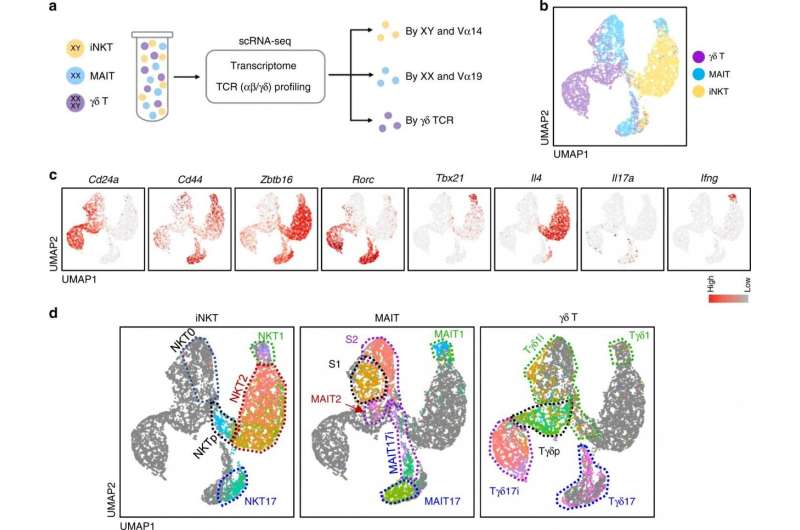
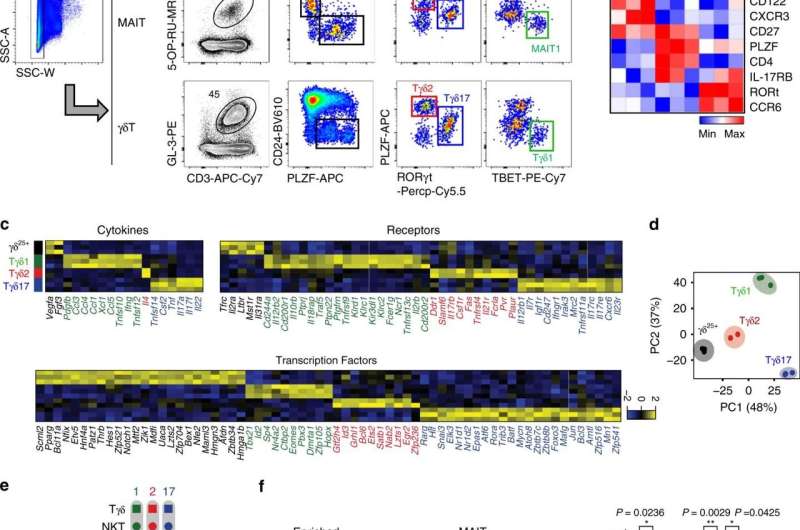
The recently discovered innate T cell is made in active form from the developmental stage without having encountered the pathogen and account for 20-30% of all T cells, but its production process or role was not well known. In response, the team focused on the developmental process of three congenital T cells common in humans and mice: natural killer T cells, gamma delta T cells, and MAIT cells.
These cells, which were believed to have disparate developmental systems and functions through single cell genomic analysis, actually share the same developmental path from each precursor and has been found to differentiate into functional subtypes that secrete the same cytokines, such as interferon gamma, interleukin-4, and interleukin-17.
In examining the composition of congenital T cell subtypes, mice have many natural killer T cells, but humans have many MAIT cells or gamma-delta T cells. Because of this, strong anti-cancer and antiviral efficacy of natural killer T cells that secrete interferon gamma in mice are verified but it is difficult to expect the equal effect in humans who possess a very low number of natural killer T cells.
The study has found that MAIT cells or gamma-delta T cells in humans are functionally equivalent to the natural killer T cells in mice. The research team anticipates that immunotherapy using MAIT and gamma-delta T cells, which secrete interferon gamma in humans, will produce anti-cancer and antiviral effects as they do in mice in the future.
More information: Minji Lee et al, Single-cell RNA sequencing identifies shared differentiation paths of mouse thymic innate T cells, Nature Communications (2020). DOI: 10.1038/s41467-020-18155-8