Scientists capture 3-D images of tumors in subcellular resolution
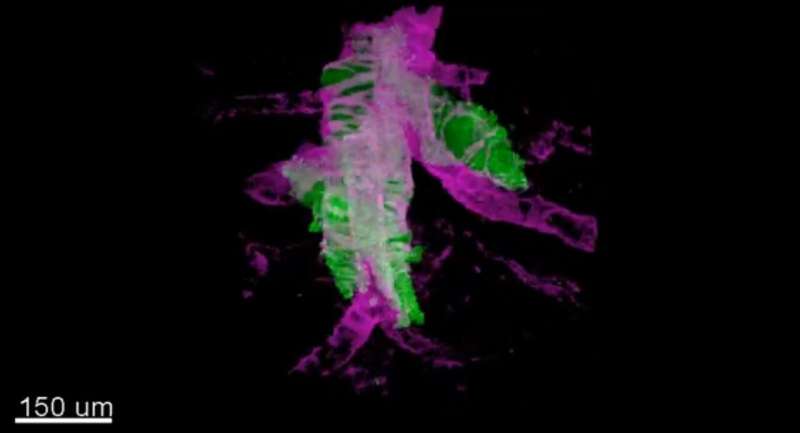
Researchers at the Francis Crick Institute have developed a new imaging technique that allows simultaneous visualization of multiple tissue types in 3-D, providing one of the most comprehensive views of conditions like cancer to date.
Details of their new method called 'fast light microscopic analysis of antibody stained whole organs', or FLASH, published in Nature Protocols today (Friday), highlight how the technique can be used to examine intact tissue and even whole organs without harm to the sample integrity.
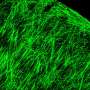
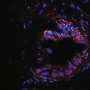
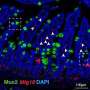
3-D FLASH of mammary epithelium showing luminal cells (pink), myoepithelial cells (yellow) and basal lamina defining adipocytes
Seeing the big picture
The research team set out to address problems with existing histology techniques that prevent a truly holistic view of diseases like cancer. Most imaging methods require ultra-thin samples of just a few micrometers thick, which may not be reflective of the rest of the sample and cannot provide insight into the spatial interactions between different cell types. For example, how cancer cells and immune cells interact.
Over a period of about two years, they trialed different combinations of reagents, techniques and timings, until they managed to find a process that allowed them to capture the detailed 3-D images, while leaving the sample or organ intact for further analysis.
Hendrik Messal, first author and postdoctoral fellow at the Netherlands Cancer Institute, said: "The concept of 3-D imagery is fast evolving and researchers are always testing the boundaries of what we can see using the latest technologies and techniques.
"FLASH is a fast and easy method that can be quickly adopted by researchers familiar with light microscopy techniques, and importantly, it doesn't damage the tissue which allows for additional diagnostic tests."
Guiding cancer treatment
FLASH is already showing promise for guiding future clinical decisions. In a letter in Nature, published last year, the team showed how the technique could be used to examine the architecture of pancreatic tumors and predict how aggressive the cancer would become.
"Thin histological sections might miss some clues of cancer biology," says Jorge Almagro, first author and Ph.D. student in the Crick's Adult Stem Cell Laboratory.
"Examining a whole sample, provides researchers with a panoramic view of disease—which cell types are present and how they are interacting."
The research team are continuing to refine this method and adjust it to suit different tissue types and research questions.
Axel Behrens, group leader of the Crick's Adult Stem Cell Laboratory, and new director of the Cancer Research UK Convergence Science Centre at Imperial Collage and the Institute of Cancer Research, said, "This technique is an excellent example of the value of multidisciplinary science. It's in applying knowledge of chemical and physical principles to medical research questions, that we are able to accelerate the development of new technologies and make a difference for patients."
More information: Hendrik A. Messal et al. Antigen retrieval and clearing for whole-organ immunofluorescence by FLASH, Nature Protocols (2020). DOI: 10.1038/s41596-020-00414-z