Mutations in key chromosome protein may lead to neurodevelopmental issues
Researchers from Children's Hospital of Philadelphia (CHOP) have demonstrated that newly mutated variants of a key protein involved in supporting chromosomes result in several issues ranging from developmental delays to epilepsy to congenital abnormalities and may provide researchers with a new target for therapeutic intervention. The findings were published online today by the journal Science Advances.
Histones are proteins that allow chromosomes to achieve their compacted shape while still passing on vital genetic information. The histone H3.3 has been extensively studied in its involvement in the development of cancer. However, the mutations that have been studied are somatic mutations, or mutations that occur in a single cell of the body and cannot be inherited. However, there is very little research on how H3.3 is impacted by germline mutations, or mutations that effect every cell in the body.
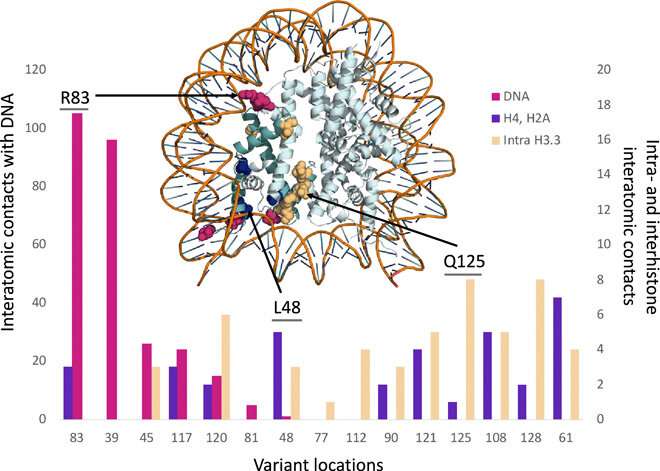
To determine whether germline mutations of the H3.3 histone had any impact on patients, the study team analyzed 46 patients with 37 different germline mutations who also had neurologic dysfunction and congenital anomalies without malignancies. Using molecular modeling, the researchers identified that all of 37 variants demonstrated clear disruptions in interactions with DNA, other histones, or histone chaperone proteins. The effect that these germline mutations had on patients was clearly distinct from the effect of somatic mutations in the same histones.
"By analyzing these variants, we identified a novel genetic syndrome associated with a variety of neurological impacts, including neurodegeneration," said Elizabeth Bhoj, MD, Ph.D., an attending physician with the Division of Human Genetics at CHOP and senior author of the study. "With more work, we may be able to develop targeted therapies for patients affected by these mutations."
More information: Laura Bryant et al. Histone H3.3 beyond cancer: Germline mutations in Histone 3 Family 3A and 3B cause a previously unidentified neurodegenerative disorder in 46 patients, Science Advances (2020). DOI: 10.1126/sciadv.abc9207