Neutrons reveal unpredicted binding between SARS-CoV-2, hepatitis C antiviral drug
Scientists have found new, unexpected behaviors when SARS-CoV-2—the virus that causes COVID-19—encounters drugs known as inhibitors, which bind to certain components of the virus and block its ability to reproduce.
Published in the Journal of Medicinal Chemistry, the research provides key insights for advancing drug design and drug repurposing efforts to treat COVID-19.
Researchers at the Department of Energy's Oak Ridge National Laboratory used neutron scattering to investigate interactions between telaprevir, a drug used to treat hepatitis C viral infection, and the SARS-CoV-2 main protease, the enzyme responsible for enabling the virus to reproduce.
They discovered unforeseen changes in the electric charges in the drug binding site of the protease enzyme that were not predicted by prevailing computer simulations.
"We found this particular protein—the SARS-CoV-2 main protease—which a lot of researchers are studying by computational methods, is behaving in a surprising way," said lead author Daniel Kneller. "Our findings provide critical experimental data needed to improve computer modeling so that simulations more closely match reality."
The virus reproduces by assembling long chains of proteins, or polyproteins, made from almost 2,000 amino acids that must be cut into smaller chains by the main protease. Finding a drug that effectively blocks or inhibits the protease function is paramount to preventing the virus from replicating and spreading to other healthy cells in the body.
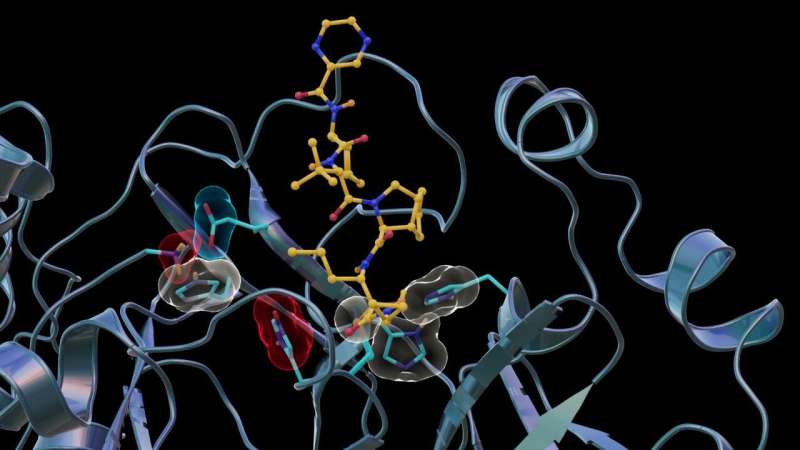
In previous studies, the team used neutron and X-ray scattering to build a complete map of every atom in the SARS-CoV-2 protease enzyme. They located the sites where a drug inhibitor would bind to the protease enzyme and mapped the network of hydrogen bonds that hold the protease together. They also determined the locations of the positive, negative and neutral electric charges in the protease amino acid sites where the polyprotein cutting action occurs.
"In this study, we discovered that when telaprevir bound to the main protease, the protonation states [proton addition or loss] in the binding site altered, meaning the locations of the hydrogen atoms changed; essentially, some amino acid sites either gained or lost hydrogen atoms, which changed their electric charges," said ORNL corresponding author Andrey Kovalevsky.
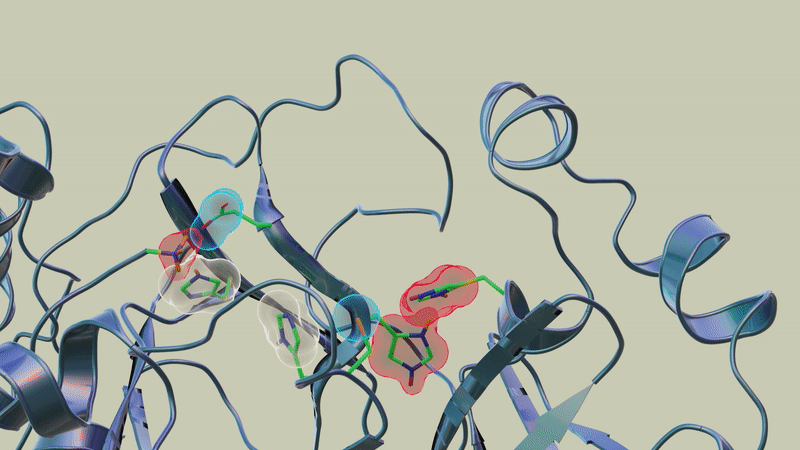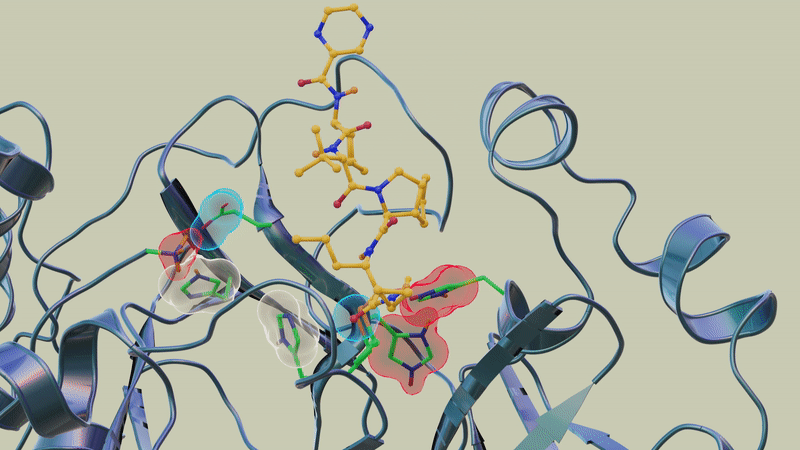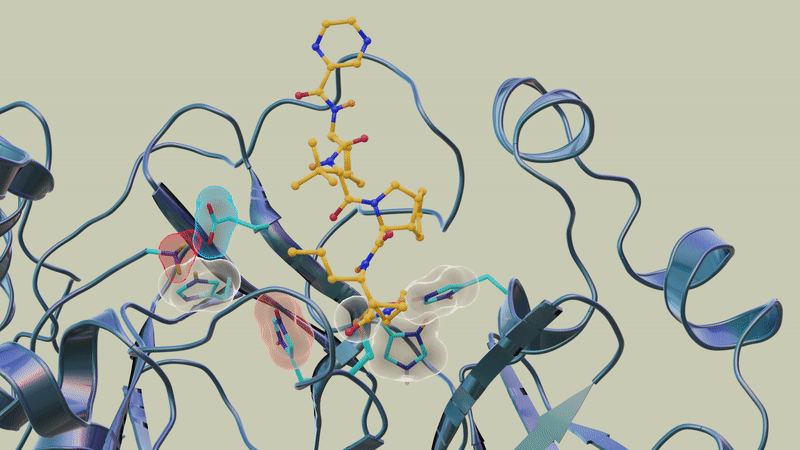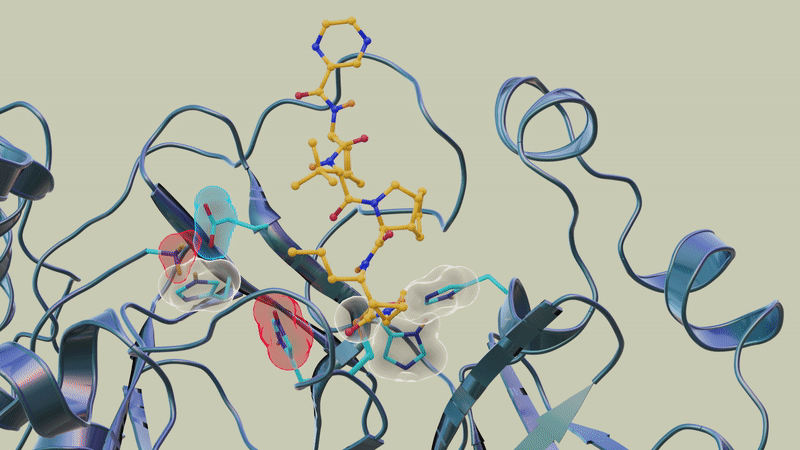
"However, the overall electric charge of the protease inhibitor binding site remained the same as it was before it was attached to the drug molecule. That's something we didn't expect and wasn't correctly predicted in computer simulations."
The researchers said their observations suggest the protease enzyme can alter its protonation states when it binds to certain drugs, and therefore, assumptions about binding behaviors should not be based purely on the properties of the protease structure before an inhibitor is bound.
"Protein behavior at the level of individual atoms is notoriously difficult to predict. Simulations have to be designed based off ideal scenarios of general chemical knowledge and mathematics, but proteins don't always adhere to ideal scenarios," Kneller said.
Telaprevir is one of a number of FDA-approved drug inhibitors used to treat hepatitis C viral infection. It belongs to a class of agents called covalent peptidomimetic inhibitors that work by using strings of unnatural amino acids to bind to specific target proteins.
These inhibitors also include boceprevir and narlaprevir—two additional hepatitis C antiviral drugs the ORNL researchers studied in 2020 as potential drug repurposing candidates to treat COVID-19.
"We've shown that some of the hepatitis C virus protease inhibitors can also inhibit the SARS-CoV-2 main protease," said ORNL's Leighton Coates, the paper's co-corresponding author.
"In this study, we chose telaprevir because we were able to produce high-quality crystal samples that we could study with neutrons. This should have a direct impact on drug design.
"We believe what we learn from the interactions between telaprevir and the protease enzyme should be transferable to other covalent peptidomimetic inhibitors such as those being investigated right now by major pharmaceutical companies," Coates added.
Neutron scattering is the primary tool for scientific discovery in the team's ongoing research campaign into the SARS-CoV-2 main protease. Neutrons are uniquely suited to detect hydrogen atoms. They have no electric charge and are nondestructive, making them ideal probes for biological studies conducted at near-physiological temperature, such as this work.
More information: Daniel W. Kneller et al, Direct Observation of Protonation State Modulation in SARS-CoV-2 Main Protease upon Inhibitor Binding with Neutron Crystallography, Journal of Medicinal Chemistry (2021). DOI: 10.1021/acs.jmedchem.1c00058