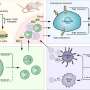
Using high-resolution, single-cell profiling to understand immune response in severe COVID-19
A new study by Yale researchers adds to our knowledge of how the body's immune system fights the virus that causes COVID-19.
The exaggerated immune responses characterizing severe COVID-19 infection are not well understood. In response to injury or infection, the immune system exhibits a coordinated response between its different components that eliminates the infection without harming the infected person. Yale researchers discovered that in patients with severe COVID-19, this coordination is lost, leading to uncontrolled amplification of the immune system.
The research was published Jan. 21 in Nature Communications.
They made their discoveries using blood cells obtained from patients throughout disease progression, at early and late points of time in the disease in patients who were hospitalized but recovered, and in those with progressive COVID-19 who required hospitalization and eventually succumbed to the disease.
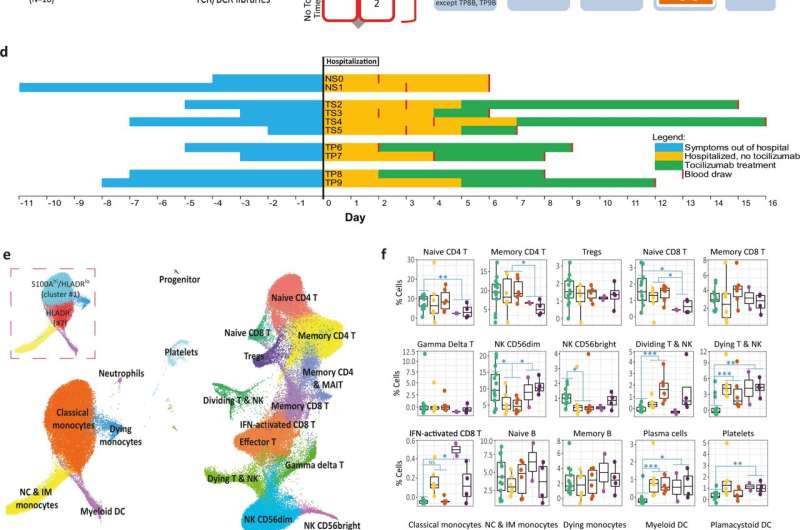
The researchers applied novel technologies called single-cell multi-omics, that dramatically increase the resolution of molecular analysis and allow characterization of all of the genes, and many of the proteins, in every single cell. "This allows accurate definition of the immune cells, the pathways and mechanisms that are activated in them, and the signals they send to other cells," explained the paper's co-first author, Avraham Unterman, MD, MBA, formerly of Yale's Kaminski Lab who is currently the head of the pulmonary fibrosis service at the Pulmonary Institute at Tel Aviv Medical Center. Naftali Kaminski, MD, is the Boehringer Ingelheim Pharmaceuticals, Inc. and Professor of Medicine at Yale School of Medicine and a senior author on the study. The technique is called single-cell multi-omic technologies.
The adaptive immune system, which attacks pathogens, and the innate immune system, which initially recognizes the infection, are supposed to be coordinated, added Charles Dela Cruz, MD, Ph.D., also a senior author on the paper. "When they are not coordinated, the clearance of the virus is delayed, the infection is amplified, and the host immune response to the virus becomes dysregulated, which can be very dangerous for patients."
The paper's co-first author, Tomokazu S. Sumida, MD, Ph.D., assistant professor (neurology), added, "This was an amazing effort. We collected the samples, and applied the most novel technologies to profile the immune system with results that provide true insight into the disease process driven by the virus." David A. Hafler, MD, William S. and Lois Stiles Edgerly Professor of Neurology and Professor of Immunobiology is a senior author on the study.
"To develop therapies for severe COVID-19, we need to understand the immune response," said Dela Cruz. "While there are now anti-inflammatory interventions, they are not specific. The findings from this study may allow identification of new novel therapies. This work was really a testament of the collaborative efforts of many individuals who responded to the pandemic." Clinical samples were obtained through the efforts of the Yale IMPACT team who recruited patients with COVID-19.
Having an interdisciplinary research team was a key to the study's success, said contributor Xiting Yan, Ph.D. "The fusion created when immunologists, physicians, and biologists work with computational biologists and bioinformaticians is very powerful and was essential for the study," Yan said.
More information: Avraham Unterman et al, Single-cell multi-omics reveals dyssynchrony of the innate and adaptive immune system in progressive COVID-19, Nature Communications (2022). DOI: 10.1038/s41467-021-27716-4