Automated assessment of pathology image quality
The analysis of tissue samples for cancer diagnosis and treatment is still largely done under the light microscope. But researchers are now developing technologies to speed up and ultimately improve the accuracy of such diagnostics through the digitization and computer-assisted analysis of tissue biopsy images. These new technologies rely a great deal on artificial intelligence (AI) tools, which require the development and "training" of AI algorithms on large datasets of digitized whole slide images (WSIs) linked to clinical outcomes data. But images collated from multiple diagnostic laboratories can vary drastically in their quality, which can in turn compromise the training and subsequent performance of AI algorithms.
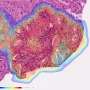
A new publication in Scientific Reports led by Ludwig Oxford's Jens Rittscher and his Oxford colleague Maryam Haghighat describes an artificial intelligence tool named PathProfiler that automates the quality control of large retrospective pathology image datasets to increase their usability in downstream research.
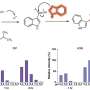
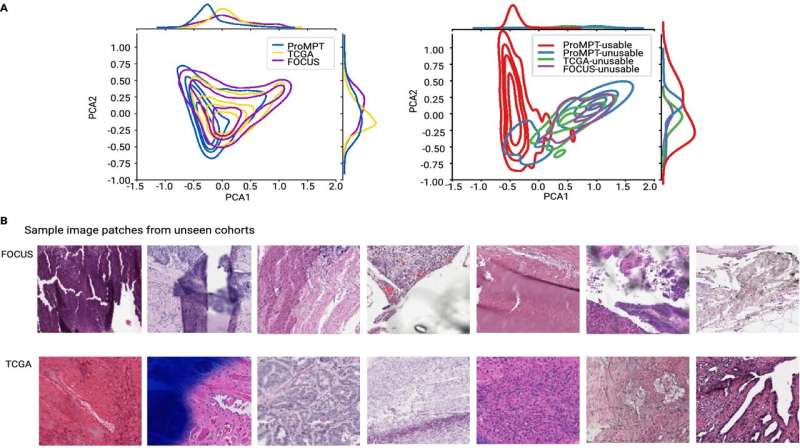
Rittscher and his Oxford colleagues developed PathProfiler using retrospective pathology images from the ProMPT (Prostate Cancer Mechanisms of Progression and Treatment) prostate cancer cohort. Their AI tool automates quality assessment of pathology images and identifies a range of potential image artifacts. It also assigns a usability score to each WSI, which will help to guide whether an image can be included in the AI training dataset. The scores generated by the AI algorithm and those assigned by three expert pathologists were highly correlated (0.89).
To further test PathProfiler, the team assessed the Cancer Genome Atlas prostate and FOCUS colorectal cancer cohorts. In addition to providing a quality score and identifying quality-impacting artifacts in the WSIs, PathProfiler was also able to predict which images could be improved by, for example, re-scanning or re-staining. This prediction is of particular relevance to the usability of samples from highly curated retrospective cohorts, such as those used in prostate cancer research.
The PathProfiler software is available to the public so that other groups can use it for their own research and contribute to its further development. The team now plans to further optimize the model using other tissue types and cohorts, and to assess the performance and utility of the tool within a clinical pathology digital pipeline.
More information: Maryam Haghighat et al, Automated quality assessment of large digitised histology cohorts by artificial intelligence, Scientific Reports (2022). DOI: 10.1038/s41598-022-08351-5