New method can easily identify viruses in tumors in a routine clinical genomic sequencing assay
Researchers have developed a method to accurately detect viruses from clinical next-generation sequencing and describe novel associations between specific tumors and viruses that warrant further investigation. This information makes it more feasible to consider viral status in treatment protocols. Their findings appear in the Journal of Molecular Diagnostics.
Viruses commonly transfer from normal human cells to malignant cells in solid tumors. However, virus detection is limited in current clinical practice. Universal screening of viruses is not technically or fiscally viable because current standard of care techniques are uniplex, costly, and have challenging workflows.
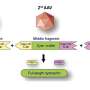
Researchers developed a digital subtraction technique, which deletes human genome cells from sequencing analysis to identify the presence of viral DNA as a quality assurance (QA) process. This bioinformatic technique requires no additional sequencing, and virus detection can be achieved with minimal additional cost. Their results were comparable to standard clinical methods for tumor virus identification.
"We decided to look at tumor types that are commonly associated with a virus, and in almost all cases, the QA tool detected the virus we expected," explained lead investigator Chad M. Vanderbilt, MD, Department of Pathology, and Department of Laboratory Medicine, Memorial Sloan Kettering Cancer Center, New York, NY, U.S.. "With this encouraging finding, we decided to refine the method as a microbiome detection pipeline and extend the analysis to see how well the method works for detecting clinically relevant viruses and discovering unexpected virus-tumor relationships."
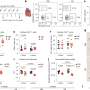
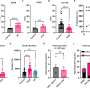
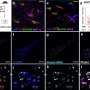
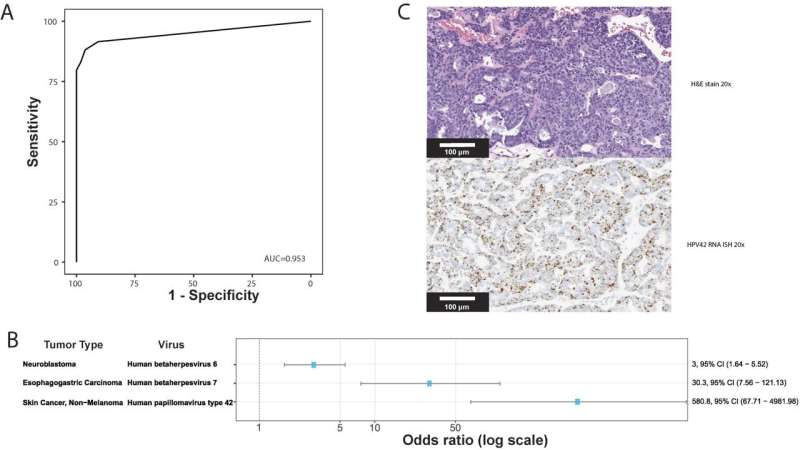
The study is the largest and most comprehensive study of human DNA virus detection in cancer. It used data gathered from January 2014 to October 2020 from 48,148 solid tumors sequenced by a US Food and Drug Administration–cleared tumor profiling assay for patients with advanced solid tumors. A BLAST (basic local alignment search tool) algorithm compared the non-human (unmapped) sequencing reads present in the tumors against all human viruses from the National Center for Biotechnology Information Virus database. Researchers cross-validated their results with multiple methods across tumor types and virus species and found their method has comparable sensitivity for detecting high risk human papilloma viruses (HPV) and Epstein Barre virus (EBV) to clinically validated in situ hybridization and amplification methods.
Investigators then extended the analysis to discover novel tumor-virus associations. Previously unreported associations between human herpes virus (HHV)6 in neuroblastoma and HHV7 in esophagogastric cancer were validated using an independent dataset. They also found a new association between HPV42 and digital papillary adenocarcinoma. In comparison to performing laborious single virus discovery assays, having access to data for discovery by data analysis alone allows resources to be dedicated to investigating the role that viruses might play in oncogenesis and for consideration of virus-informed therapies.
"The findings of this project further support the relevance of studying the role of the microbiome in disease, while the method used in this study is portable to smaller laboratories," said co-investigator Anita S. Bowman, MS, Department of Pathology, Memorial Sloan Kettering Cancer Center, New York, NY, U.S. "In the fast-paced world of oncology, effectively identifying these tumor-virus relationships adds to the collective knowledgebase and may also lead to improved therapeutic options. Ultimately, understanding tumorigenesis after a viral infection will help to attain our shared goal of saving lives."
More information: Chad M. Vanderbilt et al, Defining Novel DNA Virus-Tumor Associations and Genomic Correlates Using Prospective Clinical Tumor/Normal Matched Sequencing Data, The Journal of Molecular Diagnostics (2022). DOI: 10.1016/j.jmoldx.2022.01.011