Researchers identify genes that maintain healthy neurons
Scientists at the Francis Crick Institute have found that deleting two genes that encode key enzyme proteins (kinases NDR1 and NDR2), impairs the health of neurons and leads to neurodegeneration in young mice as well as in adults.
Their study of mouse neurons highlights the essential role of these proteins in maintaining brain health and preventing disease, a finding that could help with the discovery of future treatments for neurodegenerative diseases like Parkinson's and Amyotrophic lateral sclerosis (ALS).
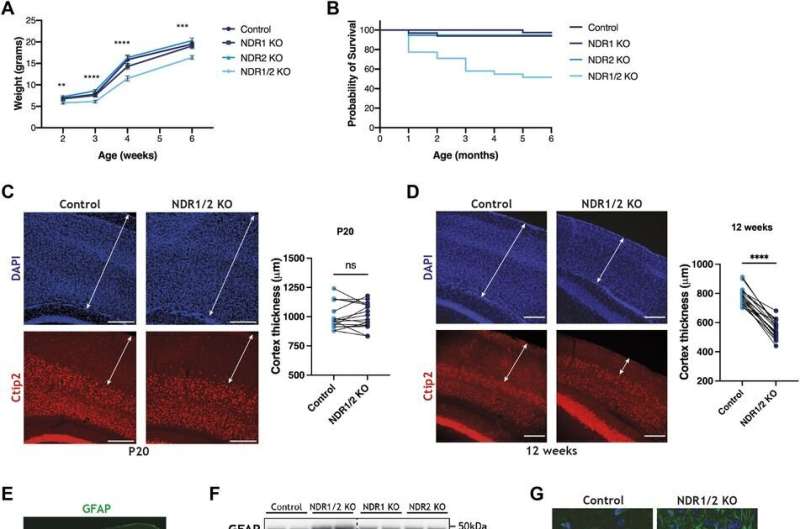
As part of their work published in Life Science Alliance on November 29, the researchers set out to understand the role of kinase enzymes in the development of the nervous system and maintaining healthy neurons. For the first time in mice, they deleted genes that encode for both kinases NDR1 and NDR2 in neurons.
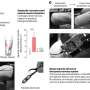
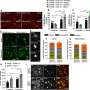
They found that deleting of either of the enzymes alone had no effect on neuronal health, but when both were eliminated simultaneously, the loss caused neurodegeneration.
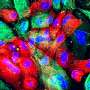
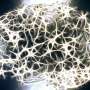
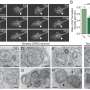
In order to understand why neurodegeneration occurs in the absence of these enzymes, the team further analyzed the brain tissue and found accumulation of protein clusters tagged for removal, a key characteristic of many neurodegenerative diseases. This suggests that the kinase enzymes are essential for the neurons to perform autophagy, the process of removing old or damaged components.
"The neuron's ability to remove toxic proteins is an essential defense against neurodegeneration," says Sila Utanir, head of the Crick's Kinases and Brain Development Laboratory.
"Understanding that kinases NDR1 and NDR2 are vital to autophagy is important because if there was a way to boost their activity with future medicines, it could help clear the protein accumulation associated with disease."
The team also looked in even more detail at the mechanisms at play when these key enzymes are lost. They found that ATG9A, a protein found in some cellular membranes, which is associated with autophagy and lipid recycling, was incorrectly positioned and as such, couldn't function properly.
Flavia Roșianu, first author of the paper, said, "The complex signals sent between cells in our brain are all part of a bigger picture of neuronal health.
"In order to understand how our brain develops and why disease occurs, we need to piece together these connections and identify the most significant proteins and signals."
More information: Flavia Roşianu et al, Loss of NDR1/2 kinases impairs endomembrane trafficking and autophagy leading to neurodegeneration, Life Science Alliance (2022). DOI: 10.26508/lsa.202201712