This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Creating an artificial pathologist
A team from the Max Planck Institute for the Science of Light (MPL) in Erlangen has created a new, fast and precise method for clinicians to analyze cells in tissue samples from cancer patients without the need for a trained pathologist. They use artificial intelligence to evaluate the data their method produces.
During cancer surgery, fast and accurate information about the operated tissue is required to guide the surgeon's next steps. When operating on a cancer patient with a solid tumor, the surgeon will send a biopsy sample to a pathologist for a rapid assessment. The pathologist needs to evaluate, for example, whether the tissue is healthy or not or how far the cancer has spread into the organs. The conventional process of such intraoperative diagnosis is time-, resource- and labor-intensive.
But what if there was a method that could do this analysis of solid tumors in only 30 minutes, accurately and in the absence of a trained pathologist? That is precisely the result of the effort of scientists from MPL and the Max-Planck-Zentrum für Physik und Medizin (MPZPM), in cooperation with the Friedrich-Alexander-Universität Erlangen-Nürnberg, the University Hospital Erlangen and the Fraunhofer Institute for Process Automation (IPA) in Mannheim.
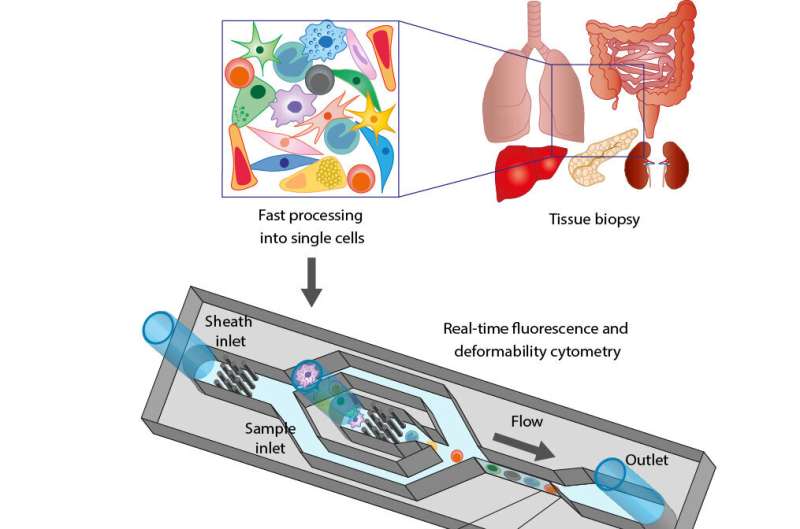
In a study published in Nature Biomedical Engineering, the team around Dr. Despina Soteriou, Dr. Markéta Kubánková and MPL director Prof. Jochen Guck use a tissue grinder, developed by IPA, to quickly tear apart biopsy samples down to the single-cell level.
These single cells are then analyzed using real-time deformability cytometry (RT-DC), a label-free method developed in the Guck laboratory. With it, they can analyze the physical properties of up to 1,000 cells per second. This is 36-thousand times faster than older, more "traditional" methods for analyzing cell deformability.
A physics approach to cell analysis
In RT-DC, single cells are pushed at high speed through a microscopic channel, where they deform under the stress and pressure. An image is taken of each cell.
From the images, scientists determine various physical properties of the cells, such as their size, shape or deformability. Kubánková explains the benefit of focusing on the physical attributes of cells: "When you go to your doctor, he or she doesn't just look at you, but also does a physical examination and feels parts of your body. With traditional methods to analyze a biopsy sample, a pathologist can only look at cells. We can do the physical examination of the individual cells, and that gives us much more information to work with."
Simply doing the physical examination of cells is not enough for diagnostics. Physicians must be able to evaluate these results without the help of a trained pathologist or physicist. In order to achieve this, the MPL group combined the tissue grinder and RT-DC with a third tool: artificial intelligence (AI).
The AI model evaluates the large complex datasets obtained by RT-DC analysis and quickly assesses whether a biopsy sample contains tumor tissue or not. The use of AI also affirmed the importance of cell deformability as a biomarker, as the results were notably worse when the AI was not trained with this variable. Overall, the entire procedure including sample processing and automated data analysis takes less than 30 minutes and is therefore fast enough to be done during surgery.
Importantly, a pathologist does not need to be readily available to evaluate the sample. This is a great advantage, because intraoperative consultations are not always possible and at some sites, samples can only be analyzed after the surgery is concluded. Depending on the result this often means that days later, the patient has to return to the hospital for another surgery.
In addition to testing for the presence of tumors, the method was also used to detect tissue inflammation in a model of inflammatory bowel disease (IBD). In the future the method could help clinicians to assess disease severity or distinguish between different types of IBD.
The team aims to one day, transfer their method to a clinical setting to aid or even replace the classical pathological analysis. Soteriou is happy with the results of their study: "This was a proof of concept study—the method could accurately determine the presence of tumor tissue in our samples very quickly. The next step will be to continue to work very closely with clinicians to determine how this method can best be translated into the clinic.
MPL director, and current speaker of the MPZPM, Prof. Guck comments, "This is a first major success of the new MPZPM, and a prime example of how new physical approaches can contribute to advancing long-standing problems in clinical medicine, such as accurate cancer diagnosis. With the completion of the MPZPM building on the medical campus next summer, the essential interactions between physicists and clinical researchers will become even more frequent and substantial, and more such breakthrough results can be expected."
More information: Jochen Guck, Rapid single-cell physical phenotyping of mechanically dissociated tissue biopsies, Nature Biomedical Engineering (2023). DOI: 10.1038/s41551-023-01015-3. www.nature.com/articles/s41551-023-01015-3