April 13, 2023 report
This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Reduced editing implicated in mitochondrial cascade of schizophrenia related RNA
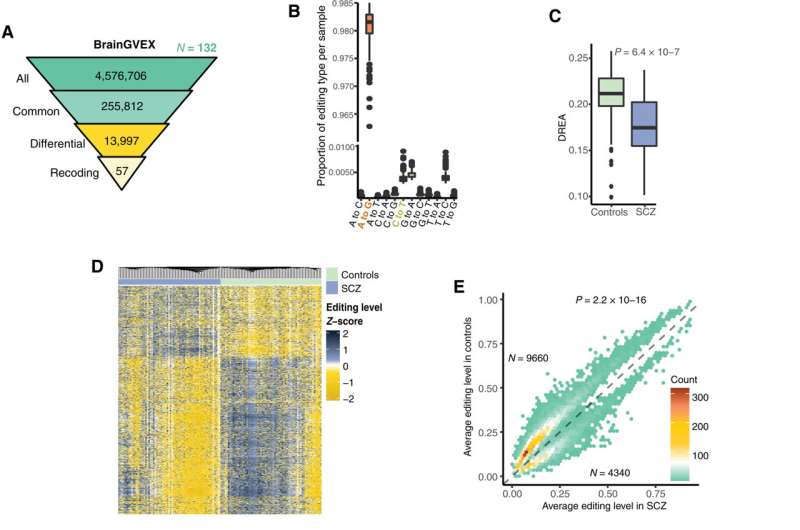
Researchers at the University of California at Los Angeles have analyzed RNA editing in postmortem brains of four schizophrenia cohorts and uncovered a significant and reproducible trend of hypo-editing in patients of European descent.
The paper "Widespread RNA hypo-editing in schizophrenia and its relevance to mitochondrial function," published in Science Advances, details the research team's efforts to isolate functionally impacting RNA editing sites to understand how dysregulated editing contributes to various disorders.
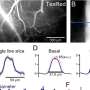
In the data analysis, researchers identified 26,841 unique differential editing sites. They observed a significant trend of lower than expected amounts of RNA editing in the schizophrenia groups, which was reproduced in three of the four cohorts of European individuals.
The common trend of less edited RNA in individuals with schizophrenia across multiple cohorts revolved around genes involved in brain-related pathways such as motor neuron axon guidance, synaptic transmission, and ion transport.
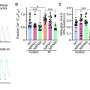
According to the researchers, the global reduction of RNA editing seen in these schizophrenia diagnosed brains implies the deregulation of myriad biological processes. For that to happen, there must be disrupted mechanisms of editing. The findings support the association between enzyme ADAR3 and reduction of editing, as there was a significant negative correlation between overall editing levels and ADAR3 gene expression.
Previous studies have proposed that ADAR3 negatively regulates neuronal differentiation by regulating mRNA stability and translation. Studies have also correlated lower levels of ADAR3 with malignant increases in pathological gliomas, highlighting the need to understand the complex roles of the enzyme's regulatory activities.
However, ADAR3 does not regulate how ADAR3 is produced. That responsibility falls on elements within the mitochondria. The current study observed mitochondria with severely fragmented phenotypes. Mitochondrial fusion gene MFN1 was being affected by the under-editing, and this gene encodes a mitochondrial membrane protein essential for mitochondrial fusion. With fusion inhibited, the natural mechanism of mitochondria repair is impacted, which can result in altered transcription levels of RNA.
The current study highlights a growing field within science of understanding the editing, splicing and post-translational modification processes that operate on an otherwise static genome. The amount of translated RNA and untranslated regulatory RNA encoded in the genome is sparse in comparison to the myriad of modified products required for cellular functions throughout the body.
While the study found some likely related pathways and proteins correlated to RNA editing suppression, future work is required to further investigate the regulatory mechanisms of reduced editing.
Further challenges await as the European and African American cohorts showed the complete opposite trend of editing changes. While the European cohorts showed consistent under-edited RNA, the African American cohorts showed a hyper-editing bias related to schizophrenia. This suggests that different mechanistic alterations to RNA editing regulation can lead to schizophrenia in diverse ancestral populations, which could have consequential disparities in patient diagnosis and treatment.
More information: Mudra Choudhury et al, Widespread RNA hypoediting in schizophrenia and its relevance to mitochondrial function, Science Advances (2023). DOI: 10.1126/sciadv.ade9997
© 2023 Science X Network