This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
New method enables detection of plasma biomarkers cheaply and at scale
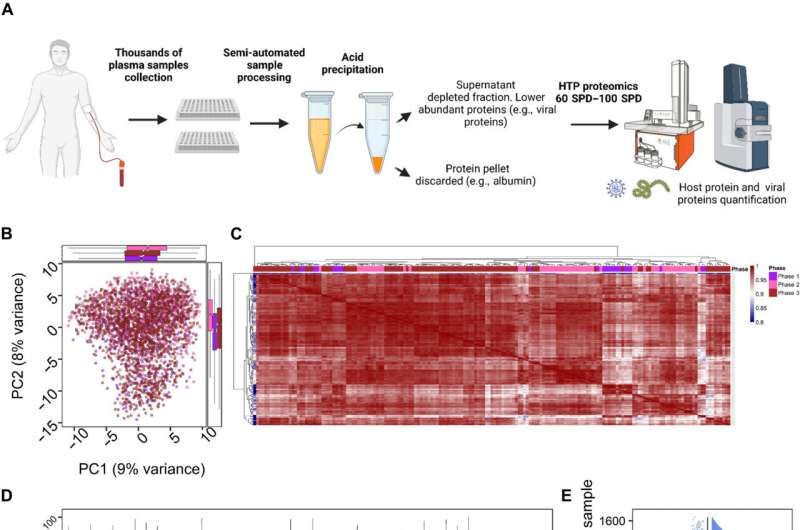
Blood plasma is collected from people routinely during clinical care and for research. It is potentially a rich source of protein biomarkers for diagnostic and prognostic purposes, for measuring response to treatment, and for revealing disease biology. Yet identifying such biomarkers in plasma with proteomics, our best available tool, has been a challenge: 99% of plasma consists of everyday proteins like albumin, globulins, and coagulants that essentially crowd out the needle-in-a-haystack proteins that could serve as biomarkers.
"Plasma is a notoriously difficult body fluid when it comes to proteomics," says Hanno Steen, Ph.D., director of Boston Children's Proteomics Center. "You detect the same proteins again and again in a sample, so it is difficult to go deep enough to detect the interesting, less-abundant proteins."
People had been using antibodies to eliminate the common proteins from plasma samples, but the method is expensive, slow, and not robust enough to be used in thousands of samples. In a new report in Science Advances, Steen and colleagues describe a method that removes these proteins in a rapid, cost-effective manner—tinkering with an old biochemical method dating back to 1948.
They had seen the technique described in the literature from time to time in other studies of proteins and peptides and realized it could be applied to plasma. "We then spent time optimizing and streamlining the process so that it works in 96-well plates on thousands of samples," says Steen.
Putting the strategy to the test: COVID-19
Necessity was the mother of invention. Steen and his team needed a method to analyze the results of the IMPACC study, a large national study funded by the National Institute of Allergy and Infectious Diseases (NIAID), in which Boston Children's has played a key role. The study is examining immune responses in about 1,100 people hospitalized with COVID-19.
"We were tasked by NIAID to analyze the proteins in 5,000 plasma samples," says Steen. "At a time when many labs had to shut down due to the pandemic, we had three months to develop a method to analyze 10 times more plasma samples every day than we had ever done. Nobody had ever done deep plasma protein proteomics at this scale. I said yes, even though I had no idea how to do it."
The team ably rose to the occasion, and their work helped reveal tissue- and organ-specific biomarkers that accurately predicted the course of hospital patients with COVID-19; that work is expected to be published later this year.
Precipitating proteins out
The technique uses perchloric acid, an extremely strong acid also called a superacid, combined with very low temperatures (-20°C) to precipitate the common proteins out in as little as 15 minutes, without damaging the other proteins.
"The unwanted proteins coagulate like milk in lemon juice," says Steen. "You can immediately see them curdling. You then can centrifuge the sample and take the liquid at the top—that has the proteins we want."
The technique is fast enough to use at scale, depleting the unwanted proteins at a cost of just $2.50 per sample—far below current costs—and enabling detection of more than 1,500 proteins per sample at a rate of 60 samples per day.
"Working under time pressure probably helped," says Steen. "We were under the gun and had to put a method together."
More information: Arthur Viode et al, A simple, time- and cost-effective, high-throughput depletion strategy for deep plasma proteomics, Science Advances (2023). DOI: 10.1126/sciadv.adf9717