This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Computational method uncovers the effects of mutations in the noncoding genome
Less than two percent of the human genome codes for proteins, with the rest being noncoding and likely helping with gene regulation. Mutations in the noncoding genome often trigger trait changes that cause disease or disability by altering gene expression. However, it can be hard for scientists to track down which of numerous variants associated with a disease or other complex trait are the causal ones and to understand the mechanism of their effects.
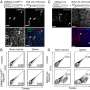
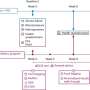
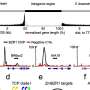
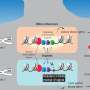
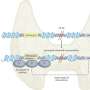
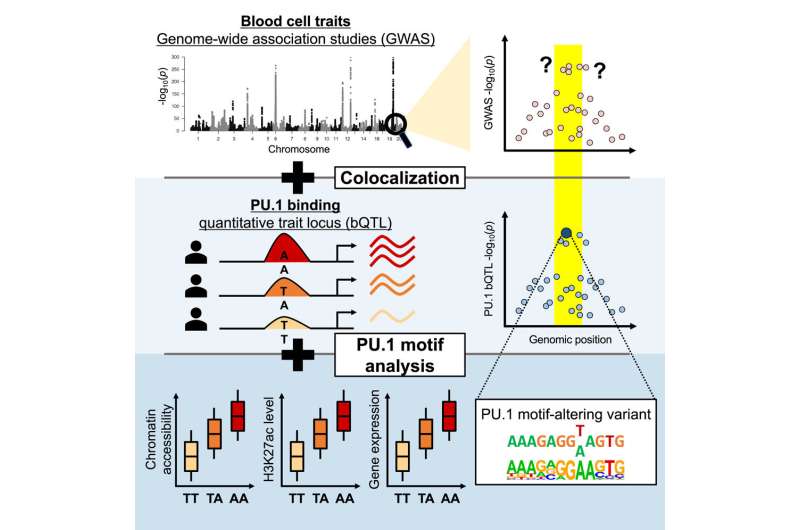
Researchers at the Brigham developed a new computational approach that hones in on small regions of the noncoding genome that genome-wide association studies (GWAS) identified as being correlated with changes to blood cell traits, including lowered lymphocyte counts and hemoglobin concentrations.
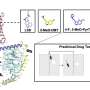
They then scanned these regions for specific mutations that caused a transcription factor protein, called PU.1, to bind to certain areas more or less strongly than normal, and examined the effect that such mutations had on PU.1's binding site. Their method uncovered 69 mutations that affected PU.1 binding and were related to quantitative differences in blood cell trait changes, 51 of which altered PU.1's binding site and thus likely caused a physiological difference.
"Our method could be applied to better understand a range of genetic conditions and to help pinpoint the causal variants in the noncoding genome underlying various biomedical traits," said senior author Martha Bulyk, Ph.D., a Principal Investigator in the Brigham's Division of Genetics.
"Here, we identified noncoding variants that appear to contribute to quantitative differences in blood cell trait changes. This approach could be used to uncover the transcriptional regulatory mechanisms hidden in the GWAS data of other complex traits."
The study is published in the journal Cell Genomics.
More information: Raehoon Jeong et al, Blood cell traits' GWAS loci colocalization with variation in PU.1 genomic occupancy prioritizes causal noncoding regulatory variants, Cell Genomics (2023). DOI: 10.1016/j.xgen.2023.100327