This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
A new method for evaluating the pathogenicity of missense variants using AlphaFold2
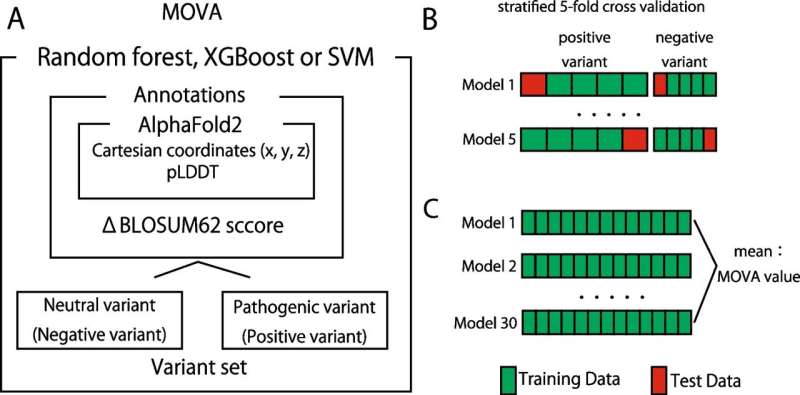
The Department of Neurology at Niigata University has developed a new in silico method for evaluating the pathogenicity of missense variants using AlphaFold2 (MOVA).
Rare variants in the causative gene of ALS are present in 10 to 30% of sporadic ALS cases, which highlights the need for accurate and efficient pathogenicity prediction methods. To predict the pathogenicity of the variants, in silico analysis methods are commonly used. In some ALS causal genes, the mutations are concentrated in specific regions, and the accuracy of pathogenicity prediction can be improved by considering the positional information of the variants.
However, existing methods have not considered information on the position of variants in the protein's structure. MOVA was developed to address this issue and focuses on using positional information in the 3D structure to evaluate the pathogenicity of missense variants. The use of a machine learning method, random forest, in the development of MOVA has also shown promising results.
"The comparison of MOVA with existing in silico analysis methods, such as PolyPhen-2, CADD, REVEL, EVE, and AlphScore, demonstrates its potential in pathogenicity prediction. Combining MOVA with existing methods, such as REVEL and CADD, further improves performance beyond existing pathogenicity prediction methods alone. Moreover, MOVA also showed superior pathogenicity discrimination of hotspot mutations in the TARDBP and FUS genes. This highlights the importance of considering the positional information of variants in protein structures for improved pathogenicity prediction," says Dr. Hatano and Dr. Ishihara.
The results of the study were published in the online edition of the journal BMC Bioinformatics.
More information: Yuya Hatano et al, Accuracy of a machine learning method based on structural and locational information from AlphaFold2 for predicting the pathogenicity of TARDBP and FUS gene variants in ALS, BMC Bioinformatics (2023). DOI: 10.1186/s12859-023-05338-5