July 31, 2023 report
This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Breast cancer study counters conventional single-cell model, suggests multiple founder events common
Research led by Kyoto University, Japan, is reshaping commonly held assumptions about cancer proliferation. In their paper "Evolutionary histories of breast cancer and related clones" published in Nature, the research team explore the early evolutionary events leading to cancer development and the role of non-cancer clones that share common mutations.
The researchers used phylogenetic analysis to trace the evolution of breast cancer and precursor lesions from acquiring initial driver alterations to developing clinically diagnosed diseases.
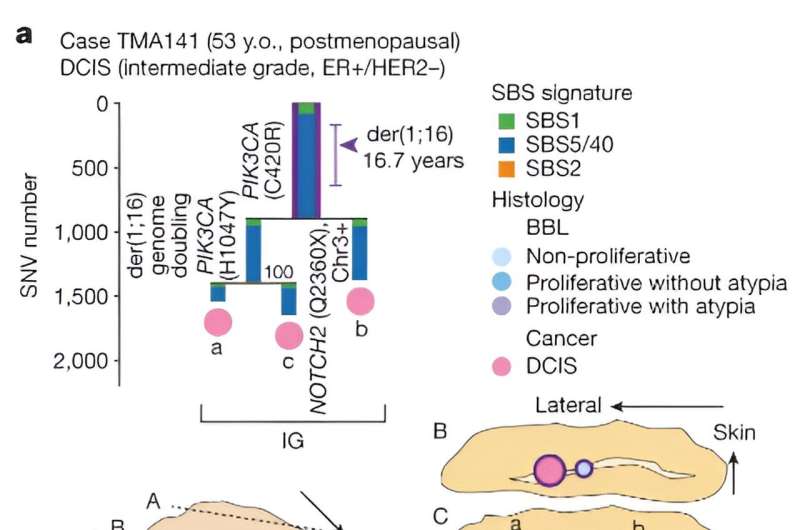
Citing recent studies that show clones carrying common cancer mutations can exist in healthy tissues, the researchers wanted to know more about the driver events and their order of occurrence before these clones evolve into cancer. The study focused on breast cancers harboring a genetic alteration called der(1;16), found in approximately 20% of breast cancers.
The researchers estimated the approximate timing of early evolutionary events based on the mutation rate measured in normal epithelial cells. They found that the acquisition of the der(1;16) genetic alteration occurred during early puberty to late adolescence. By the patient's early 30s, a common ancestor emerged, from which both cancer and non-cancer clones evolved.
The researchers performed whole-genome sequencing (WGS) of multiple samples obtained from both cancer and clonally related benign breast lesions, along with apparently normal lobules. They estimated the rate of mutation accumulation from WGS of single-cell-derived organoids established from mammary epithelia. Based on this mutation rate, they reconstructed phylogenetic trees that included both cancer and non-cancer clones to infer the entire history of breast cancer evolution.
Researchers estimated the rate of mutation accumulation in normal mammary epithelial cells as they age with 71 single-cell-derived organoids from normal mammary tissues of breast cancer patients and breastfeeding healthy volunteers. The study identified somatic mutations in these organoids and analyzed their mutation rates.
A linear regression model determined that the number of single-nucleotide variants (SNVs) significantly depended on factors such as age at sample collection, years after menopause, parity (number of pregnancies reaching 20 weeks), and the presence of a driver mutation.
Before menopause, SNVs accumulated at 19.5 mutations per genome per year, which reduced to 8.1 mutations per genome per year after menopause. For each parity event, the mutation number was reduced by 54.8, suggesting that giving birth impacts the mutation accumulation rate in mammary cells.
The high reduction of SNVs per parity seems substantially more significant than researchers expected from the typical time frame of menstrual cycle interruption by pregnancy (1.1–1.5 years). This could mean that the mammary epithelium is being reconstructed by newly recruited, "dormant" stem cells. A similar process has been proposed to explain the disappearance of clones carrying tobacco signatures in bronchial epithelium after smoking cessation.
The presence of PIK3CA mutations increased the number of SNVs by 210.4. PIK3CA mutations can cause the PI3K enzyme to become overactive, which may cause cancer cells to grow and is associated with many types of organ cancer.
Indels were also studied, and their accumulation rate was reduced by 45% after menopause, from 1.3 mutations per genome per year to 0.72 mutations per genome per year. Each delivery reduced the mutation number by 3.9.
Multiple independent cancer founders from non-cancer ancestors were common, contributing to intratumor heterogeneity. This differs from the conventional thinking that most cancers evolve from a single cancer founder, suggesting that multiple cancer founder cells from within a non-cancer population might be more common than expected.
The mutational profile of mammary epithelium is distinct from that of other tissues, more influenced by dynamic changes occurring across a woman's lifespan, such as menstrual cycles, pregnancy, delivery, and breastfeeding. The current research hints at the possibility of detecting precancerous clonal cells long before any cancer emerges.
More information: Tomomi Nishimura et al, Evolutionary histories of breast cancer and related clones, Nature (2023). DOI: 10.1038/s41586-023-06333-9
© 2023 Science X Network