This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers develop tool that could improve liquid biopsy
A research team led by Xianghong Jasmine Zhou, Ph.D., professor of Pathology and Laboratory Medicine at the David Geffen School of Medicine at UCLA, has made an important advancement to address one of the major challenges in cell-free DNA (cfDNA) testing, also known as liquid biopsy.
They've identified specific methylation patterns unique to each tissue, potentially helping to Identify the specific tissue or organ associated with cfDNA alterations picked up by testing, a critical challenge for accurate diagnosis and monitoring of diseases.
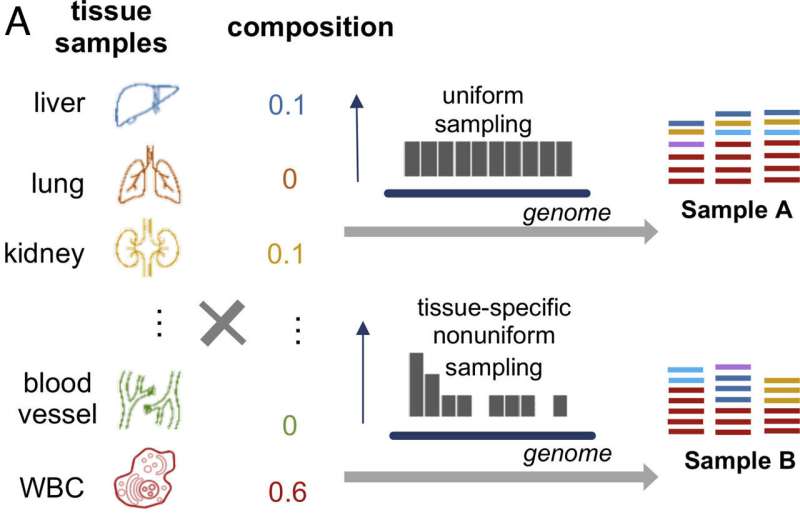
Cell-free DNA has significant potential in disease detection and monitoring. However, accurately quantifying tissue-derived cfDNA has proven challenging with current methods, among them determining the tissue origin of cfDNA fragments detected in these tests.
In a new study, published in Proceedings of the National Academy of Sciences, the team developed a comprehensive and high-resolution methylation atlas based on a vast dataset of 521 noncancerous tissue samples representing 29 major types of human tissues.
They call the approach cfSort, and showed it successfully identified specific methylation patterns unique to each tissue at the fragment level and validated these findings using additional datasets.
Going further, the team illustrated the clinical applications of cfSort through two potential uses: aiding in disease diagnosis and monitoring treatment side effects. By estimating the tissue-derived cfDNA fraction using cfSort, they were able to assess and predict clinical outcomes in patients.
"We have shown that the cfSort outperformed the existing methods in terms of accuracy and detection limit: making more accurate tissue fraction estimation and distinguishing a lower level of tissue-derived cfDNA," said first author Shuo Li.
"In addition, the cfSort demonstrated nearly perfect robustness toward the unseen local fluctuations of tissue compositions, indicating its wide applicability to diverse individuals."
More information: Shuo Li et al, Comprehensive tissue deconvolution of cell-free DNA by deep learning for disease diagnosis and monitoring, Proceedings of the National Academy of Sciences (2023). DOI: 10.1073/pnas.2305236120