This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
How the herpes virus HCMV deceives its host cells
Herpes viruses are treacherous: once you are infected, you can never get rid of the virus. This is because herpes viruses lie dormant in certain host cells in the body for a lifetime. Almost every adult unknowingly carries at least one of the nine different human herpes viruses. The virus can be reactivated due to age, stress or a weakened immune system and lead to sometimes severe diseases.
Herpes viruses are so successful because they have adapted well to humans and developed effective strategies to escape the immune system. Proteins that make the infected cell believe that it is not infected or threatened play a central role in camouflage. It is known, for example, that every herpes virus has a powerful proteome, i.e. a large number of these proteins, which, highly adapted to the host, enables it to replicate efficiently immediately after infection.
The complex proteome also ensures that multilayered particles are built up in the already infected cell. These newly formed viruses—also called virions—contain numerous viral proteins as well as host proteins. In the center of the particles is the viral DNA, which is enclosed by a nucleocapsid. A layer of numerous other proteins called tegument is formed around this capsid.
Particles come into play in the reactivation of the virus
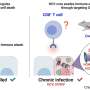
The particles are crucial in enabling the virus to replicate again and spread systemically in the body after reactivation triggered by whatever means. They are therefore central to the outbreak of disease—after a long period of dormancy (latency).
However, little is known about the internal organization of these particles, especially the protein-protein interactions within the tegument. Researchers from the Leibniz-Forschungsinstitut für Molekulare Pharmakologie (FMP) and the Charité-Universitätsmedizin Berlin have therefore taken a closer look at the particles, specifically in human cytomegalovirus (HCMV).
HCMV occurs particularly frequently in the population and can be really dangerous, especially for transplant recipients and unborn children who become infected via the mother. Despite intensive research, there is currently no well-tolerated antiviral therapy that could effectively control or even eliminate the virus. There is also no vaccination against this type of virus.
Map shows which proteins interact with each other
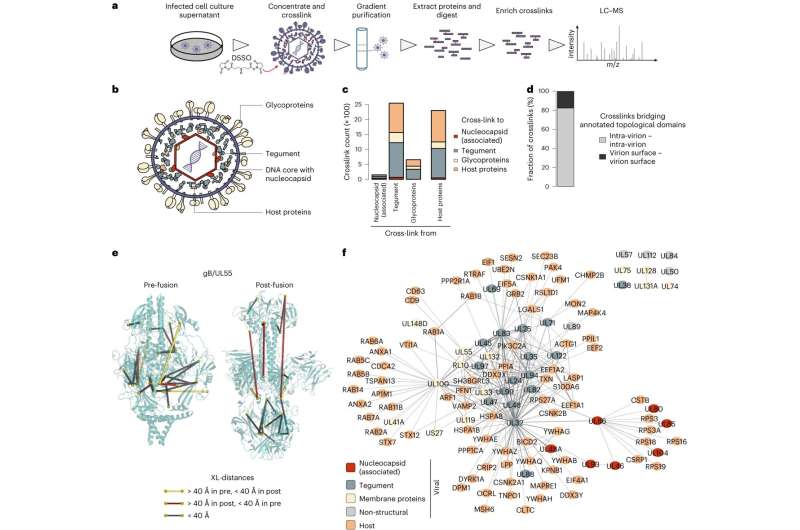
In the current work, the team led by Fan Liu (FMP) and Lüder Wiebusch (Charité) has for the first time created a detailed map of the spatial interactions between viral and host cell proteins within HCMV particles. Among other things, this revealed that certain host cell proteins are recruited by viral proteins and play a role in viral replication. For example, a viral protein called UL32 recruits a cellular protein (protein phosphatase , PP1) into the particle to avoid binding of other, unwanted, host cell proteins.
"HCMV itself does not have any phosphatases like PP1, so you can see that the virus takes advantage of certain host cell proteins to replicate efficiently," says FMP virologist Boris Bogdanow, explaining a key strategy for how HCMV tricks its host.
To study the interactions between the different proteins in intact HCMV particles layer by layer, the researchers used a technique called cross-linking mass spectrometry. "This method also allows us to draw conclusions about the identity of the proteins," emphasized Fan Liu, an expert in mass spectrometry at the FMP. "But what is special and unique about cross-linking is that we can see which proteins interact with each other and where."
Never before has this innovative technology been used to map the spatial organization of interactions within herpesviral particles. With the data thus obtained, a computer model of the HCMV particle was subsequently created at FU Berlin by Mohsen Sadeghi. The virtual model allows simulation of each protein within the particle and visualizes the biophysical processes in a vivid way.
"The identified protein-protein interaction is important to better understand the complex life cycle of HCMV," Boris Bogdanow classifies the results. "And this, in turn, is important for finding candidate anti-viral drugs against HCMV."
More information: Bogdanow, B. et al. Spatially resolved protein map of intact human cytomegalovirus virions. Nature Microbiology (2023). DOI: 10.1038/s41564-023-01433-8 www.nature.com/articles/s41564-023-01433-8