This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Interpreting large-scale medical datasets: Generative model enables multi-scale representations of cells and samples
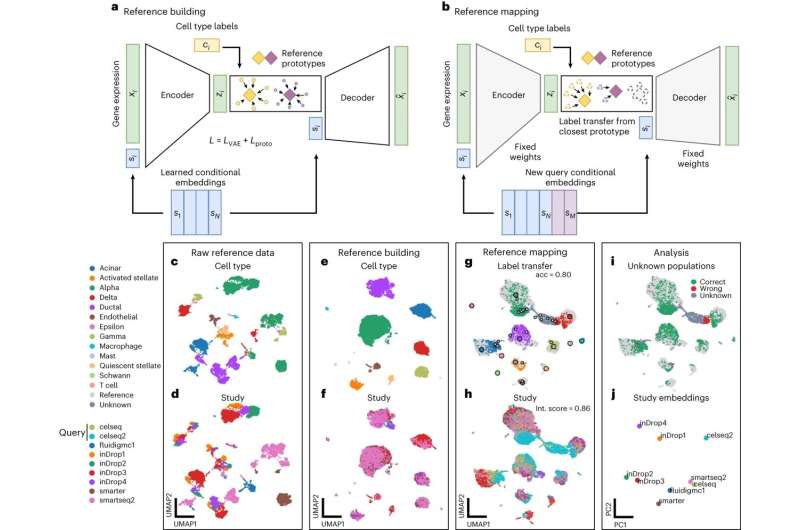
The increasing amount of data recorded in medical research can only lead to scientific breakthroughs and essential therapies for patients if interpreted and analyzed correctly. Computer scientists at Helmholtz Munich developed a generative model named scPoli (single-cell population level integration), that performs data integration of high-quality large-scale datasets of single cells to create valuable single-cell reference maps of the human body, so-called single-cell atlases, for medical research.
Atlases offer a quick and detailed approach for data interpretation, that can be applied by the medical research community, ultimately leading to novel biological insights and disease understanding. The model has now been introduced in Nature Methods.
In recent years, we have witnessed an astronomical surge in both the quantity and intricacy of data, especially in the field of medical research. Scientists are now able to capture tissues and organs in fascinating detail—at the level of single cells. Combing the resulting datasets has led to the creation of so-called single-cell atlases, which are reference maps of every cell present in a specific organ with the goal of creating a map of the entire human body.
These high-quality large-scale datasets enable not only novel biological insights into the cellular heterogeneity of certain tissues but also accelerate various steps in the analysis workflows that are usually time-consuming. Using these atlases researchers can now for instance compare organs from healthy humans with diseased ones from patients leading to valuable findings of disease development and progression.
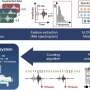
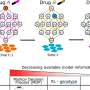
As these atlases become bigger in scale, a need for machine learning models and computational algorithms to analyze and integrate data arises. With a multi-scale representation approach of both cells and samples, which often represent patients in large-scale studies, Prof. Fabian Theis from Helmholtz Munich and professor at the Technical University of Munich, as well as Dr. Mohammad Lotfollahi and Carlo De Donno from the Computational Health Center at Helmholtz Munich have developed a new generative model named scPoli (short for single-cell population level integration).
This is the first data integration model that can produce representations for both cells and samples. With this new generative model, medical datasets can be easily analyzed to identify the main sources of variability, while at the same time being aware of the natural heterogeneity that occurs between data of different individuals and cells.
The team has developed scPoli with the intent of enhancing the interpretability of single-cell studies. Unlike other models, that only produce cell representations, scPoli offers a novel point-of-view to researchers to investigate and link patterns at the sample level which has the potential to improve integration workflows and interpretation.
The researchers at Helmholtz Munich have already demonstrated the functionality of their model by integrating data from two major single-cell atlases. The integration of the Human Lung Cell Atlas, a reference map of the lung recently published by the team around Prof. Fabian Theis showed both an improvement in performance and offered novel sample-level insight.
Furthermore, scPoli was used to integrate a large-scale PBMC atlas (a single-cell atlas of peripheral blood mononuclear cells), consisting of 7.8 million cells, which highlights the possible scaling properties, which are fundamental for large-scale integration studies.
ScPoli is unique compared to previous data integration efforts. It proposes various use cases for multi-scale analysis because, for the first time, scientists are able to simultaneously explore cell and sample or patient representations, capturing the characteristics and features of individual cells in much more detail than ever before. It is shown that scPoli can enable multi-scale classification of cells and samples, as well as guided data integration workflows.
The model has the potential to accelerate atlas building and usage, which in turn accelerates disease understanding and the development of novel therapies.
More information: De Donno et al, Population-level integration of single-cell datasets enables multi-scale analysis across samples. Nature Methods. DOI: 10.1038/s41592-023-02035-2. www.nature.com/articles/s41592-023-02035-2