This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Single-cell RNA-sequencing analysis reveals immune cell heterogeneity in five autoimmune diseases
Autoimmune diseases are a group of diseases caused by abnormal immune attacks on healthy cells, tissues or organs. Single-cell RNA-sequencing (scRNA-seq) technology provides transcriptomic information at the single-cell resolution, thus offering a new way to study autoimmune diseases. Most single-cell RNA-seq studies, however, have often focused on one type of autoimmune disease.
For this study, published in BIO Integration, scRNA-seq data was integrated from peripheral blood cells of five different autoimmune diseases (IgA nephropathy [IgAN], Kawasaki disease [KD], multiple sclerosis [MS], Sjogren's syndrome [SS], and systemic lupus erythematosus [SLE]). Dimensionality clustering, cellular communication analysis, re-clustering analysis of monocytes, NK cell populations, differential gene expression analysis, and functional enrichment was performed for all immune cells in these data.
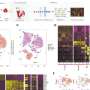
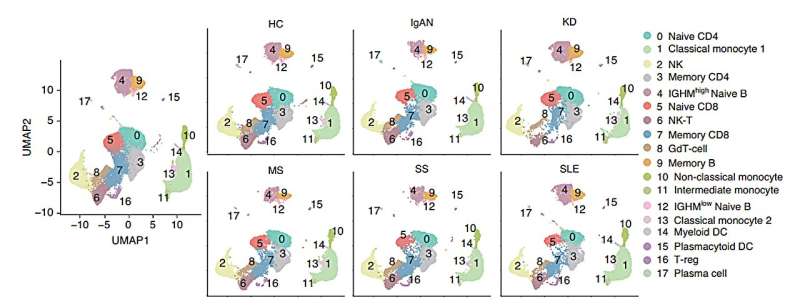
The scRNA-seq results of peripheral blood cells from five different autoimmune diseases (IgAN, KD, MS, SS, and SLE) were integrated. All samples contained 18 different immune cell subsets, although the cell cluster populations were different among the five diseases.
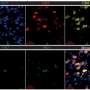
Through intercellular communication network analysis, the researchers determined that the signals of classical and non-classical monocytes were significantly enhanced in patients with IgAN and SLE. The signals of naïve B cells were increased in patients KD. Interestingly, the signals of NK and NK-T cells were enhanced in patients with SS, but reduced in patients with IgAN and SLE.
Transcriptomic analysis of classical and non-classical monocyte subsets further revealed that pro-inflammatory cytokines and interferon-related genes, including CCL3, IL1B, ISG15, and IFI6, were specifically increased in patients with IgAN and SLE. Unlike monocytes, the number and NK marker genes were decreased in patients with IgAN and KD, but increased in patients with SS. Meanwhile, two NK-T cell subsets were exclusively found in SS.
In summary, based on an integration of the single-cell RNA-seq results, the team demonstrated changes in the immune cell landscape of five different autoimmune diseases with respect to immune cell subsets, populations, differentially-expressed genes, and the cell-to-cell communication network. The data provides new insight to further explore the heterogeneity and similarity among different autoimmune diseases.
More information: Siweier Luo et al, Single-Cell RNA-Sequencing Integration Analysis Revealed Immune Cell Heterogeneity in Five Human Autoimmune Diseases, BIO Integration (2023). DOI: 10.15212/bioi-2023-0012