This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Single-cell RNA sequencing maps immune cell heterogeneity in mice with allogeneic cardiac transplantation
Immune cells play important roles in mediating allograft rejection and tolerance after cardiac transplantation. However, immune cell heterogeneity at the single-cell level, and how immune cell states shape transplantation immunity, remain incompletely characterized.
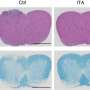
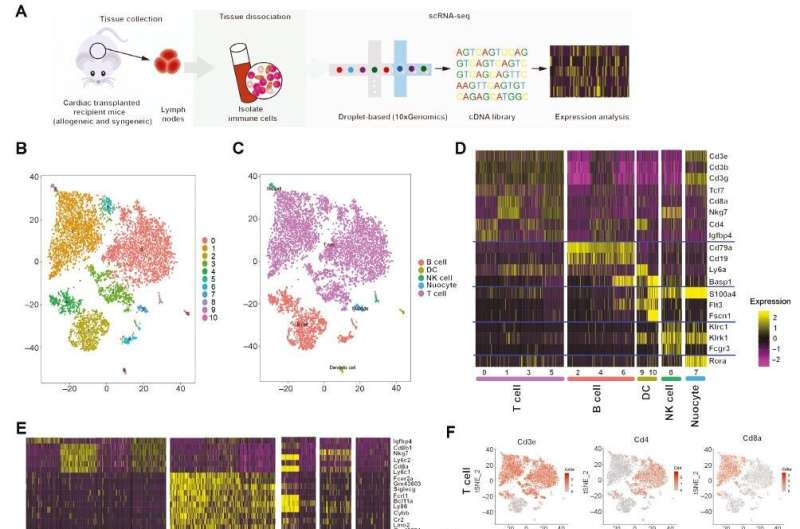
The authors of a new study, published in Cardiovascular Innovations and Applications, performed single-cell RNA sequencing (scRNA-seq) on immune cells in LNs from a mouse syngeneic and allogeneic cardiac transplantation model. Nine T cell clusters were identified through unsupervised analysis. Pathway enrichment analysis was used to explore the functional differences among cell subpopulations and to characterize the metabolic heterogeneity of T cells.
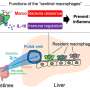
The transcriptional landscape of immune cells was determined, particularly T cells, and their metabolic transcriptomes in LNs during mouse cardiac transplantation. On the basis of molecular and functional properties, we also identified T cell types associated with transplantation-associated immune processes, including cytotoxic CD8+ T cells, activated conventional CD4+ T cells, and dysfunctional Tregs. The contribution of JunB to the induction of Th17 cell differentiation and restriction of Treg development was further elucidated, and the authors identified that HIF-1a participates in T cell metabolism and function.
The first systematic single-cell analysis of transcriptional variation within the T cell population is presented in this article, providing new insights for the development of novel therapeutic targets for allograft rejection.
More information: Zhonghua Tong et al, Single-Cell RNA Sequencing Maps Immune Cell Heterogeneity in Mice with Allogeneic Cardiac Transplantation, Cardiovascular Innovations and Applications (2023). DOI: 10.15212/CVIA.2023.0023