This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
Study validates AI diagnostic for colorectal cancer biomarker

In a study published today in Nature Communications, a team of scientists from French-American techbio Owkin and pathology labs in France present a blind validation of MSIntuit CRC, a first-in-class AI-driven digital pathology diagnostic developed by Owkin, as a pre-screening tool aimed at optimizing the precision of diagnosis and treatment of colorectal cancer.
With almost two million new cases and one million deaths worldwide in 2020, colorectal cancer is the third most common cancer globally and second highest cause of cancer mortality.
Microsatellite Instability (MSI) is a key genomic biomarker in colorectal cancer and represents ~15% of the overall CRC population. Recent clinical trials have shown that MSI phenotype has both prognostic and therapeutic importance, especially with the recent approval of immune checkpoint inhibitor (ICI) therapies.
Those patients whose tumors show MSI, are considered likely to respond to ICI therapy and recommended for it. Conversely, ICI is not routinely recommended for those with tumors that are microsatellite stable (MSS). As a result, MSI testing is now recommended by consensus guidelines across the globe as a guide to best therapy.
Prescreening tools that can remove the need to test all patients offer the opportunity to streamline the process and reduce pressure on laboratory staff and resources.
Today's study reveals that MSIntuit CRC can accurately rule out nearly 50% of MSS patients, while correctly classifying more than 96% of MSI patients, on par with current gold standard methods (92-95%). Such new solutions pave the way for an optimized screening workflow to screen more patients, faster.
"This new approach will have a direct impact on oncologist decision making and help bring the best treatment to patients sooner," states Magali Svrcek, International expert in GI pathology, Professor at Saint Antoine Hospital, Sorbonne Université, AP-HP, France and co-last author of this publication. "It could also optimize costs and organization of MSI testing in pathology labs, especially for countries applying universal MSI screening."
Meriem Sefta, Chief Diagnostic Officer at Owkin, said, "With the increasing number of biomarkers to be routinely tested in clinical practice, the need for tools that can both ease bottleneck and resource pressures while ramping up biomarker testing is paramount."
"Our solution represents the first step towards the development of an AI diagnostic that can identify actionable biomarkers from a single H&E slide used in clinical routine, pushing us closer to realizing a precision medicine future."
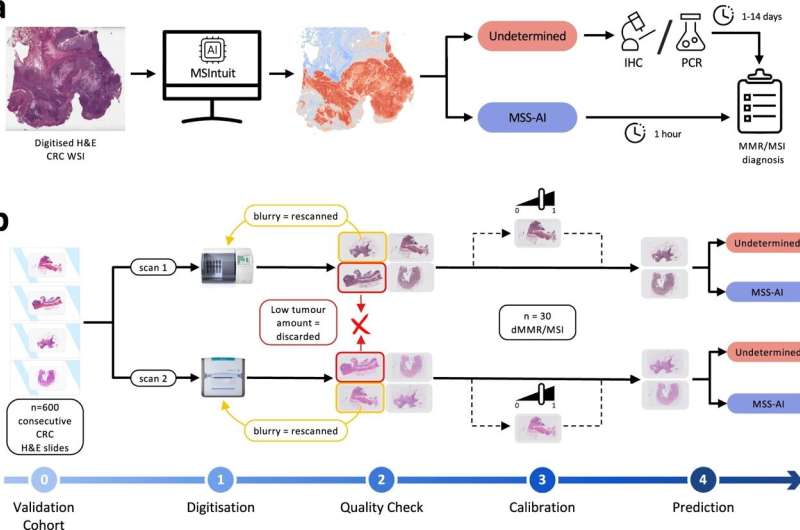
A major strength of the study is the blind validation of the model on 600 consecutive CRC cases diagnosed across nine different pathology labs in the span of two years, thus reducing risk of selection bias, thanks to Medipath, the largest pathology lab network in France. This validation was also carried out across two different pathology slide scanners and performed consistently with a sensitivity of 96% and 98% respectively.
This and other methodological approaches, such as the emphasis on sensitivity and specificity as performance markers, demonstrate a commitment to robustness of the AI model, all with the intention of ensuring the diagnostic is fit for clinical settings and generalizable across labs and geographies.
More information: Charlie Saillard et al, Validation of MSIntuit as an AI-based pre-screening tool for MSI detection from colorectal cancer histology slides, Nature Communications (2023). DOI: 10.1038/s41467-023-42453-6