This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Study demonstrates benefit of precision medicine based on race/ethnicity
"Precision medicine," an emerging approach to health care in which a patient's genetics, diseased tissue and other factors guide clinicians to personalize treatment strategies, can dramatically improve health outcomes for people with various diseases, notably cancer.
Years of analysis of cancer specimens makes such targeted treatment possible, yet the samples being studied in the United States and Europe overwhelmingly come from people who are white. That reality means innovative new cancer treatments may have missed genetic predispositions toward cancer that are pronounced in people from non-white races and ethnicities.
A new study published by researchers at the University of Oklahoma College of Medicine at OU Health Sciences highlights racial disparities in a subset of cancer patients and, using advanced biomedical research technology provides evidence for those differences at the molecular level.
The collaborative study among researchers in Oklahoma and Alabama was led by Hiroshi Yamada, Ph.D., and Chinthalapally V. Rao, Ph.D., assistant professor of research and professor in the Department of Medicine, Section of Hematology-Oncology, as well as Upender Manne, Ph.D., professor of anatomic pathology at the University of Alabama in Birmingham. The study was published in npj Precision Oncology.
"People from various races, regions, and communities face differences in the prevalence of cancer and survival rates from cancer," Yamada said.
"Poor cancer outcomes have reasons. However, the reasons have not been understood at the molecular level. Unlike social factors such as poverty or access to advanced cancer care, molecular differences in cancer can be addressed by a drug or other personalized treatment. But because molecular analyses on cancers historically have focused on samples from white people, that has created a blind spot in treatment development."
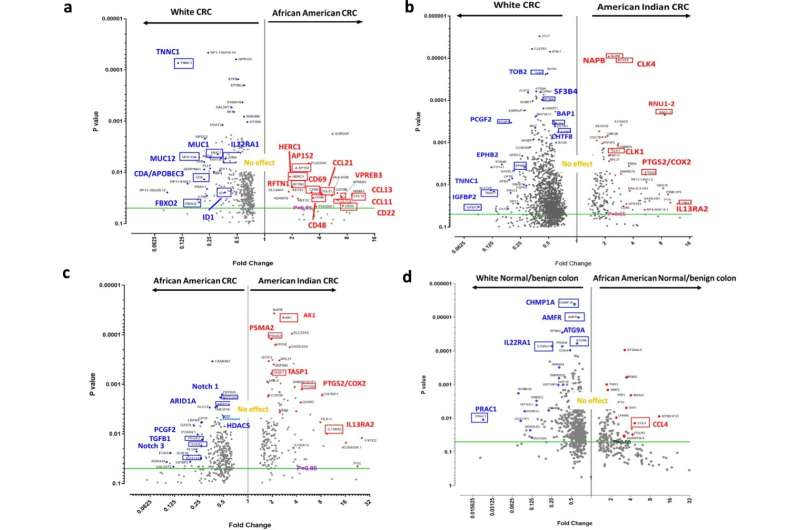
To investigate whether cancer racial disparities can be identified at the molecular level, Yamada and his team analyzed colon cancer specimens from American Indian patients in Oklahoma. In addition, researchers at the University of Alabama provided cancer specimens from African American patients in Alabama.
For many years, statistics have shown that non-white populations often face cancer at a higher rate and have worse health outcomes than people who are white. Advancements in technology now allow researchers to find evidence for those disparities at the molecular level.
Yamada and his team indeed discovered differences in American Indians and African Americans, as compared to white Americans, in two important areas: gene expression (instructions in the DNA that tell cells what to do) and cytokines (proteins that play a role in how cells interact and communicate with each other).
Because of what researchers know about those differences, some of the colon cancer drugs under development—primarily based on data from cancer samples of white patients—may be less effective for American Indians or African Americans, Yamada said.
"This study highlights potential targets for colon cancer prevention and treatment in American Indians and African Americans, which is information that would not be available through the analysis of cancer specimens from white populations alone," Yamada said.
Yamada said that racial/ethnic groups share biological "patterning" that occurs because of social constructs (not unchangeable characteristics), as well as dietary habits, geographic locations, and other cultural practices and lifestyles. It is essentially impossible to separate these elements that make up biological patterning, he said, but from the standpoint of research, race is an important trait to consider.
Yamada said he hopes his study raises awareness about the shortage of cancer specimens from non-white populations and bolsters the effort to diversify samples for research. He also plans to continue his research in this area by investigating the function of genes that could be influencing colon cancer development and treatment outcomes in American Indians and African Americans.
In the United States, colon cancer is the third most common cancer diagnosed in both men and women and, in 2022, was the second most common cause of cancer death.
"Molecular analysis is a new research tool in the field of cancer disparities," said Yamada, who is among the early adopters of the approach. "Molecular analysis according to race, or any population vulnerable to cancer, will help to identify specific cancer traits, which can be used in the development of precision medicine. This approach will improve poor health outcomes in minorities who have cancer."
More information: Hiroshi Y. Yamada et al, Molecular disparities in colorectal cancers of White Americans, Alabama African Americans, and Oklahoma American Indians, npj Precision Oncology (2023). DOI: 10.1038/s41698-023-00433-5