This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Evolutionary conservation of CD4 and LAG-3 and their cytoplasmic tail motifs with opposing immune functions
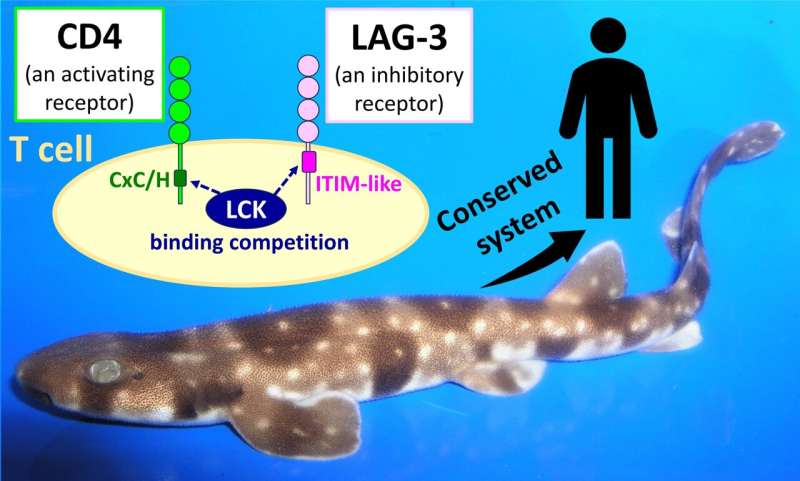
T cell surface markers CD4 and LAG-3 are related proteins that promote and inhibit cell activity, respectively. However, although LAG-3 has started to gain appreciation as an immune checkpoint molecule that can be targeted in cancer immunotherapy, only CD4 is well understood.
Now, a comprehensive study on CD4/LAG-3 family evolution by an international group of scientists, led by the Japanese researchers Dr. Fumio Takizawa of Fukui Prefectural University and Dr. Johannes M. Dijkstra of Fujita Health University, has concluded that CD4, as well as LAG-3, are conserved throughout jawed vertebrate species, disproving previous claims that sharks do not possess CD4.
The study also found the conserved functional motifs in the cytoplasmic tails of CD4 and LAG-3 are a CxC/H motif for cell activation and an ITIM-like motif for cell inhibition, respectively, supporting an emerging model that CD4 and LAG-3 have opposing functions through competitive binding of the kinase LCK.
The study by Takizawa and colleagues, published in Frontiers in Immunology, together with previous studies by others, appears to bring researchers closer to understanding the critical role and mechanism of LAG-3 in the immune system of humans and other vertebrates.
Cluster of differentiation 4 (CD4) is a famous marker molecule on the surface of helper T lymphocytes, where it forms a complex with T cell receptor chains α and β (TCRαβ) and CD3. This complex can be stimulated by binding peptide-loaded major histocompatibility complex (MHC) class II on, for example, B lymphocytes or dendritic cells. This induces T cell activation by promoting the binding of the intracellular kinase LCK to the CD4 cytoplasmic tail motif CxC/H (single letter amino acid code), which initiates an activation cascade through phosphorylation of CD3 cytoplasmic tails.
Lymphocyte activation gene-3 (LAG-3, CD223) is closely related to CD4 and also binds MHC class II, and, in the genomes of many species, CD4 and LAG-3 genes are tandemly organized. However, compared to CD4 (188,519 search matches in the PubMed database), LAG-3 has only been poorly studied (2,637 search matches).
The reason for this difference is that, unlike CD4, LAG-3 is not a convenient marker of a major immune cell lineage but, predominantly, an activation marker that is expressed on activated CD4+ or CD8+ T cells (CD8 is a marker of cytotoxic T cells) as a kind of safety brake. Only recently, the interest in LAG-3 has increased because, like PD-1, PD-L1, and CTLA-4, it is an immune checkpoint molecule that can be targeted by antibodies during cancer immunotherapy to enhance immune responses.
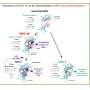
Arguably, only after studies in 2019 and 2022, following earlier conflicting information on the functional domains in the LAG-3 cytoplasmic tail, a first plausible model for the inhibitory function of mammalian LAG-3 cytoplasmic tail appears to be emerging. This model entails that an FxxL (single letter amino acid code) motif and the tail's C-terminal acidic stretch inhibit the association of LCK with CD4 or CD8 and, thereby, T cell activation.
The model implies that LAG-3 lures LCK away from CD4 or CD8 by (i) binding LCK with its FxxL motif that resembles an immunoreceptor tyrosine-based inhibition motif (ITIM) motif, known to bind SH2 domains as found in LCK, and (ii) attracting, by its acidic stretch, Zn2+ ions that are necessary for binding of LCK to CD4 or CD8 through a "zinc-clasp" that involves a CxC or CxH motif in the CD4 or CD8 cytoplasmic tail.
Dr. Takizawa and Dr. Dijkstra, as well as several of their co-authors, have >10 years of experience in studying CD4 in fish. For the present study, they embarked on the ambitious task of making a comprehensive analysis of the evolution of the CD4/LAG-3 molecule family in jawed vertebrate species.
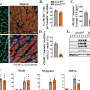
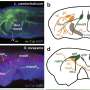
Jawed vertebrates, with sharks as the most primitive representatives, have very similar immune systems, but an earlier study claimed that sharks lack CD4 [6]. However, supported by finding conserved sequence motifs and genomic locations, Takizawa and coworkers now show that sharks do possess CD4 and also LAG-3.
They also investigated the CD4 and LAG-3 situation in bichir and sturgeon, showing that these primitive ray-finned fishes possess genes of both lineages CD4-1 and CD4-2, which was previously only known in teleost (modern bony) fish and concluded a CD4 gene duplication early in ray-finned fish evolution.
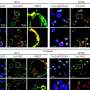
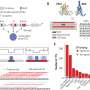
Many analyses by Takizawa and coworkers were based on the investigation of public databases, but in collaboration with the Aqua Department of the veterinary pharma firm HIPRA in Spain, the University of Maryland in the U.S., and Aqua World Ibaraki Prefectural Oarai Aquarium in Japan, they also experimentally analyzed CD4 and LAG-3 in sterlet sturgeon, nurse shark, and cloudy catshark, respectively. The expression patterns of shark CD4 and LAG-3 genes in tissues and cells were found to be reminiscent of what is known in mammals.
Previously, it was argued that a CxH motif, as found in the cytoplasmic tail of shark CD4, may not bind LCK. However, that argument disagrees with CxH motifs known to be functional in other zinc-clasps, and with evidence that the CxH motif in bony fish CD8α can form a zinc-clasp with LCK. Takizawa and coworkers demonstrate that the evolutionary change of the ancestral CxH motif to CxC in CD8α only occurred within an ancestor of lungfish and tetrapod species.
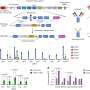
The most important findings by Takizawa and coworkers are that CD4 and LAG-3 are conserved throughout jawed vertebrates including sharks, and that they retained their cytoplasmic tail motifs CxC/H and F/YxxL in the cases of CD4 and LAG-3, respectively. In contrast, the mammalian LAG-3 cytoplasmic tail C-terminal acidic stretch appears not to be evolutionary ancient.
The F/YxxL motif is similar to an ITIM, and some fishes even have a perfect canonical ITIM motif at this site in the LAG-3 cytoplasmic tail as Dr. Takizawa and Dr. Dijkstra already realized in 2010.
In summary, the comprehensive evolutionary study by Takizawa and coworkers strongly supports that a conserved core of T cell regulation is formed by the competition between LAG-3 and CD4 or CD8 for binding LCK through an ITIM-like and a CxC/H motif, respectively. This simple model appears to be the most robust explanation for all observations and may help to finally settle the debate on how LAG-3 functions.
More information: Fumio Takizawa et al, CD4 and LAG-3 from sharks to humans: related molecules with motifs for opposing functions, Frontiers in Immunology (2023). DOI: 10.3389/fimmu.2023.1267743