This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Research identifies hantavirus in South Korea using a new rapid test, paving the way for early outbreak control
Orthohantaviruses, highly transmissible zoonotic pathogens, are notorious for causing hemorrhagic fever with renal syndrome (HFRS) in Eurasia and hantavirus cardiopulmonary syndrome in the Americas. With a significant impact on public health, they have been extensively researched for effective outbreak control and intervention strategies.
Gyeonggi Province in South Korea has reported a significant number of HFRS cases, making it a critical area for epidemiological surveillance and understanding the genomic diversity of orthohantaviruses.
To gain insights into the prevalence, viral loads, and genetic variations of Hantaan orthohantavirus (HTNV), a team of researchers led by Prof. Jin-Won Song from the Department of Microbiology, Korea University College of Medicine, conducted a study in Gyeonggi Province.
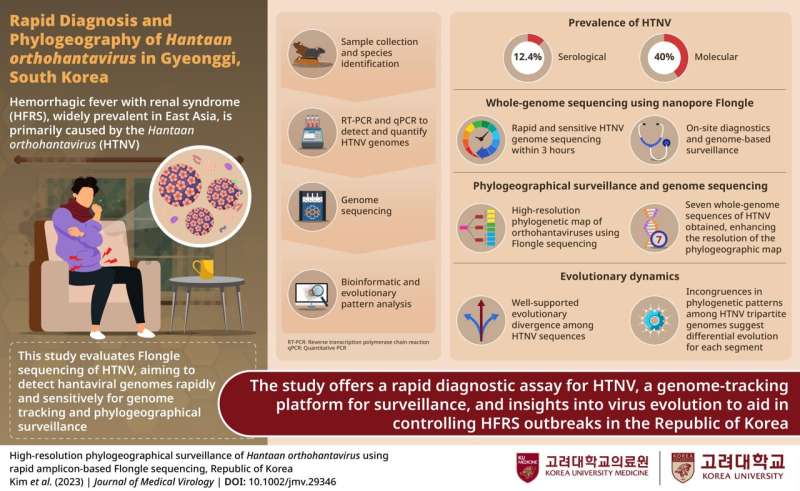
Notably, the study showcased the application of Flongle sequencing, an innovative, cost-efficient, and rapid method for detecting HTNV genomes.
"We developed a rapid and sensitive on-site diagnostic using a nanopore-based Flongle chip with a reasonable cost of around $100. This approach enables virtually whole-genome sequencing of HTNV within 3 hours," explains Prof. Song. Their work culminated in a research paper released in the Journal of Medical Virology.
The study's approach is rooted in the adoption of next-generation sequencing technologies, with a spotlight on the Oxford MinION nanopore sequencer. The comprehensive sampling strategy involved the capture of rodents and shrews from diverse regions in the ROK using live traps.
Subsequently, the team employed a multifaceted approach, incorporating mitochondrial DNA analysis, indirect immunofluorescence antibody tests, and various molecular methods for species identification and virus detection. This comprehensive methodology facilitated a thorough understanding of viral prevalence and genetic diversity.
The epidemiological surveillance conducted in Gyeonggi Province from 2017 to 2018 uncovered a significant proportion of Apodemus agrarius, a common rodent species. Notably, 12.4% of A. agrarius displayed seropositivity for HTNV, showcasing the virus's presence in the region.
Flongle sequencing proved instrumental in obtaining full-length genomic sequences from positive samples, with impressive coverage rates and accuracy comparable to Illumina sequencing. The phylogeographical analysis uncovers well-supported evolutionary divergence among HTNV tripartite genomes. Genetic clustering and incongruences in evolutionary patterns emphasize the virus's differential evolution for each genomic segment.
Prof. Song emphasizes the significance of this research by stating, "We believe our findings provide important insights into on-site diagnostics, genome-based surveillance, and the evolutionary dynamics of orthohantaviruses to mitigate hantaviral outbreaks in HFRS-endemic areas in the ROK."
"Our study pioneers the integration of cost-efficient Flongle sequencing into hantavirus diagnostics, offering a rapid and accurate tool for on-site detection. This innovation has the potential to transform how we approach and manage hantavirus outbreaks."
While the study provides groundbreaking insights, researchers acknowledge certain limitations, including the need for further sensitivity testing of Flongle-based diagnostics and more extensive genomic and epidemiological data in certain endemic areas.
This research marks a significant step forward in our understanding of orthohantaviruses, laying the foundation for more targeted control strategies against HFRS outbreaks. The findings emphasize the pivotal role of genomics in disease surveillance and offer hope for more effective responses to emerging infectious agents in the future.
More information: Jongwoo Kim et al, High‐resolution phylogeographical surveillance of Hantaan orthohantavirus using rapid amplicon‐based Flongle sequencing, Republic of Korea, Journal of Medical Virology (2024). DOI: 10.1002/jmv.29346