This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Q&A: Prescription drugs and the gut microbiome—getting the right balance
Pills and the gut microbiome sometimes don't mix. Oral prescription drugs often disturb the gut microbiome, killing off some species or changing the balance in a way that impacts patient health. In other combinations, bacteria get the upper hand and disable a drug's active ingredient.
Though the effects of many drugs on the microbiome have been documented, the mechanisms that drive these effects are mostly unknown. Understanding those mechanisms could lead to ways to counteract harmful "drug-bug" interactions or enhance beneficial ones.
Using new high-throughput genomic tools, Columbia MD/Ph.D. student Deirdre Ricaurte is now starting to reveal the ways bacteria in the gut microbiome respond to different drugs. Her research, conducted with other members of the Harris Wang lab in the Department of Systems Biology and published in the journal Nature Microbiology, could lead to improvements in precision medicine and help physicians take a patient's microbiome into account when prescribing therapy.
Ricaurte defended her Ph.D. thesis last August and is now finishing up her last two years of medical school. CUIMC News caught up with her between clinical rotations to talk about her findings and plans for the future.
What drew you to the MD/Ph.D. program at the Vagelos College of Physicians and Surgeons and microbiology research in particular?
My parents are both medical doctors, and my father has a Ph.D., so I was introduced to the MD/Ph.D. path at a pretty young age. During my first year at Princeton, I remember being captivated by a presentation that Dr. Bonnie Bassler gave to my introductory molecular biology class.
Bonnie is a brilliant researcher, renowned in the microbiology community for her contributions to the discovery of quorum sensing (the way bacteria talk to each other), and her enthusiasm for research is infectious. After that talk, I reached out to her to ask if I could rotate in her lab. My work there over the next three years became the foundation for my senior thesis.
While I entered Princeton interested in a medical career, I began seriously pursuing one after completing a global health internship with Child Family Health International. I spent my sophomore summer in Bolivia, where I worked in different rural and urban medical clinics while also volunteering in local community outreach programs.
It was a formative and inspiring experience that helped me realize my passion for medicine, particularly when practiced in underserved communities. When I was applying to medical school, I was excited about pursuing careers in science and medicine, so I decided to apply to MD/Ph.D. programs.
Beyond its stellar institutional reputation, Columbia stood out to me for its community. Its location in Washington Heights gives students the opportunity to learn from a Spanish-speaking patient population, which was very important to me. I also connected with the Columbia student body, which I think selects creative and artistic people who want to take advantage of what New York City has to offer.
Your first paper from your Ph.D. research was published earlier this year. What's the question you and your co-authors were trying to answer?
We were hoping to explore the genomic changes that bacteria go through when they are exposed to different prescription drugs and identify which of these changes might impact human treatment response. Our gut microbiome is an immense untapped source of genomic data—not just from one gut bacterium but millions of bacterial strains across hundreds of species.
To understand how prescription drugs perturb the gut microbiome, we need to access that data with next-generation sequencing technologies at an unprecedented scale. Unfortunately, many current technologies are not designed for complex bacterial communities or are too costly to apply to dozens or hundreds of bacteria at once.
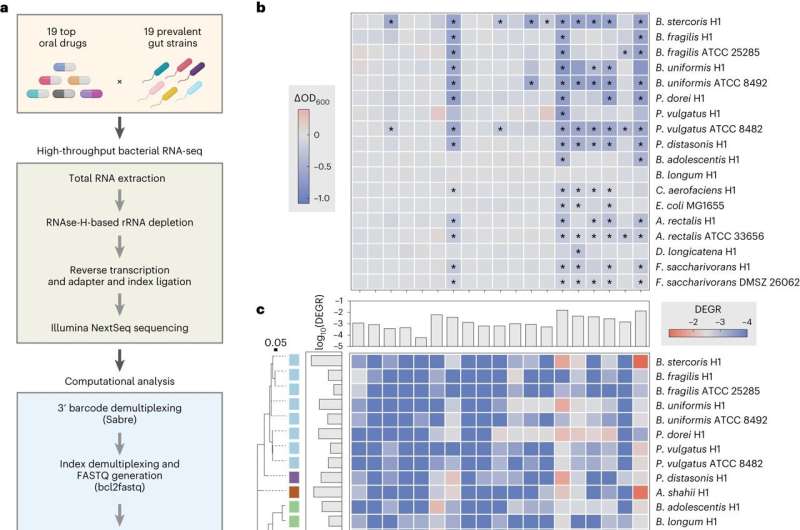
The Wang lab has made great efforts to simplify protocols and identify affordable sequencing materials. In the new study, I used a low-cost transcriptomic pipeline developed by my co-author, Dr. Yiming Huang, to examine the effects that 19 drugs had on 19 species of bacteria that live in our microbiome.
What are two of the biggest findings?
The first is that many of the drugs that we take are secret antibiotics! Unexpectedly, we found that several top-prescribed orally delivered medications are toxic to bacteria. However, toxicity against our gut microbiome is not always a bad thing. For example, our study identified how statins reduce Bacteroides numbers in the gut, a reduction that has separately been linked to lower levels of inflammation in patients taking these drugs.
Hopefully, more studies like ours will enable us to begin mapping the bacterial-drug interaction network and make more sense of the clinical responses seen in patients.
Another message is that interactions between oral medications and gut bacteria could have a powerful impact on human health and responses to treatment. Our work shows that common medications can change the way our gut bacteria respond to the vitamins and minerals we consume.
Gut bacteria may also change drug structure in ways that alter drug absorption. What's especially exciting is that the signals we see in our laboratory studies correspond to microbiome changes seen in real patient populations.
Where do you think this research will lead?
From a medical standpoint, having a scalable transcriptomic pipeline like this to assess bacterial responses to different perturbations will allow scientists to start mapping out all the possible and probable bacterial drug interactions across drug classes and different groups of patients.
Once the map is drawn, we can start thinking about ways to leverage that genetic information, like interfering with the unwanted effects of certain drugs on gut bacteria.
Cost-efficiency makes this tool more accessible to scientists globally, which will help to maximize the microbiome information generated for diverse patient populations and the clinical relevance of conclusions drawn from this information.
Once you've earned your MD, what are your plans? Will you be pursuing this line of research in the future?
I'm currently completing my clinical rotations to figure that out. I loved my OB/GYN rotation, which I just finished. There are exciting opportunities for research in women's health, and the microbiome is very relevant to that field. In fact, I have a vaginal microbiome project in the Wang Lab that is still underway.
That said, the microbiome is related to nearly every organ system in the body, and the skills I gained from the lab will serve me well in whatever field I choose. I'm excited to complete my rotations and see how a research career might fit my clinical future.
More information: Deirdre Ricaurte et al, High-throughput transcriptomics of 409 bacteria–drug pairs reveals drivers of gut microbiota perturbation, Nature Microbiology (2024). DOI: 10.1038/s41564-023-01581-x