This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Study reveals genetic clusters and biological pathways that may explain differences in type 2 diabetes risk
The development and progression of type 2 diabetes are affected by numerous biological processes, such as the body's response to insulin, the health of insulin-producing pancreatic beta cells, and the functioning of metabolic pathways.
In a recent study published in Nature Medicine that analyzed individuals from diverse backgrounds, a team led by investigators at Massachusetts General Hospital (MGH) and the Broad Institute of MIT and Harvard has identified various genetic clusters involved in a broad range of biological mechanisms that may help explain ancestry-associated differences in type 2 diabetes clinical presentations.
"Type 2 diabetes is a disease of elevated blood sugar impacting approximately one in 10 people in the United States that can cause devastating health complications and is usually not cured. It is currently not fully understood why a given person develops the disease or why there is a lot of variation around clinical features across people with the disease," says senior author Miriam S. Udler, MD, Ph.D., the director of the MGH Diabetes Genetics Clinic and an assistant professor of Medicine at Harvard Medical School.
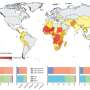
Udler and her colleagues assessed genetic findings from more than 1.4 million individuals across various genetic ancestral backgrounds: African/African American, Admixed American, East Asian, European, South Asian, and multi-ancestry.
Analyses resulted in a final set of 650 genetic variants that had independent associations with type 2 diabetes and a final list of 110 diabetes-related clinical traits.
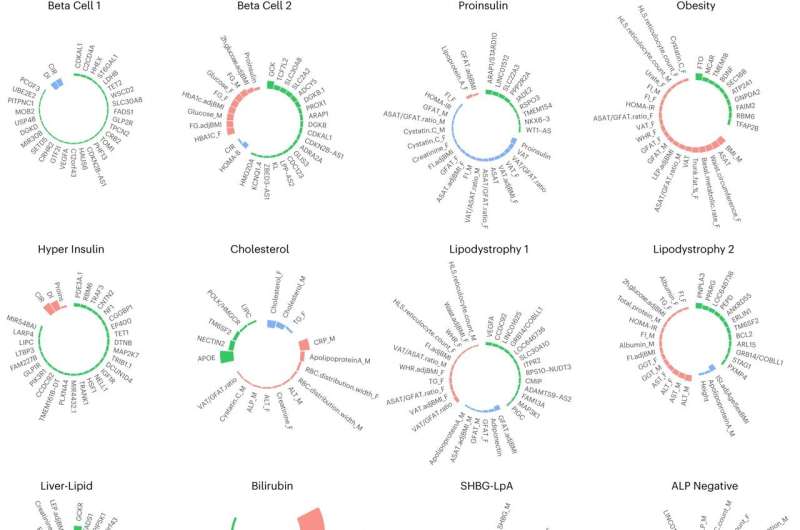
The scientists' analyses validated diabetes-associated genetic clusters that they had identified in a previous study. (A genetic cluster is a group of two or more genetic regions that are suspected to share a generalized biological function).
The work also uncovered new genetic clusters related to decreased cholesterol levels, abnormal metabolism of bilirubin (which is created when the body breaks down hemoglobin from aged red blood cells, and abnormal lipid processing in fat and liver tissues).
In addition to identifying 12 genetic clusters and their biological functions that are associated with type 2 diabetes, the investigators found that the clusters helped to explain some of the dissimilarities in type 2 diabetes among different populations.
For example, it is well-documented that individuals from various self-identified non-white populations are more susceptible to type 2 diabetes at a given body mass index.
The study's data suggest that this disparity is at least partially explained by variations in two of the clusters that the researchers identified that relate to how the body uses and stores fat—so that individuals with certain variants in these clusters (most often individuals from East Asian populations) face a higher risk of type 2 diabetes at lower body mass index levels than other individuals.
This discovery could help clinicians calculate an individual's target body mass index level based on their genetic profile.
"Our study shows that the genetic underpinnings of type 2 diabetes can help explain clinical differences across populations," says co-lead author Kirk Smith, MS, a computational biologist in MGH's Center for Genomic Medicine. "Also, the genetic mechanisms of disease that we have identified offer the potential to guide development of curative therapies," adds co-lead author Aaron J. Deutsch, MD, an instructor in the division of Endocrinology at MGH.
Additional authors include Carolyn McGrail, Hyunkyung Kim, Sarah Hsu, Alicia Huerta-Chagoya, Ravi Mandla, Philip H. Schroeder, Kenneth E. Westerman, Lukasz Szczerbinski, Timothy D. Majarian, Varinderpal Kaur, Alice Williamson, Noah Zaitlen, Melina Claussnitzer, Jose C. Florez, Alisa K. Manning, Josep M. Mercader, and Kyle J. Gaulton.
More information: Kirk Smith et al, Multi-ancestry polygenic mechanisms of type 2 diabetes, Nature Medicine (2024). DOI: 10.1038/s41591-024-02865-3