This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Study with rodents identifies key genes for control of blood pressure and heart rate
Brazilian and American researchers have identified 87 genes linked to alterations in blood pressure and 144 others associated with variations in heart rate. The findings, published in the journal Science Advances, create a unique opportunity for scientists to extend their knowledge of the origins of cardiovascular disease, the leading cause of death worldwide.
"Of the 87 genes that influence blood pressure, only 12 had been described before. The rest hadn't even been considered as possible candidates in previous studies. As for heart rate acceleration and deceleration, only 17 of the 144 genes we identified were already known. The discovery extends our knowledge of the genetic pathways that lead to regulation of blood pressure and heart rate, and will therefore help deepen our understanding of cardiovascular disease," José Eduardo Krieger, last author of the article, told Agência FAPESP.
Krieger heads the Genetics and Molecular Cardiology Laboratory at the Heart Institute (INCOR) run by the University of São Paulo's Medical School as part of its hospital complex, Hospital das Clínicas (HC-FM-USP). The study was conducted in partnership with Bruce Beutler, Director of the Center for Genetics of Host Defense at the University of Texas Southwestern in the United States.
Beutler is an immunologist and geneticist known for work on host defense mechanisms. He was awarded the Nobel Prize in Physiology or Medicine in 2011, alongside Jules Hoffmann and Ralph Steinman, for discoveries relating to the activation of innate immunity, the organism's first line of defense, especially proteins called toll-like receptors that recognize pathogens and trigger a response by the immune system.
In recent years, Beutler has developed a program at his lab that produces genotype mutations in mice for use in research on many diseases. This is now the world's largest and most advanced mutagenesis program.
Factory of discoveries
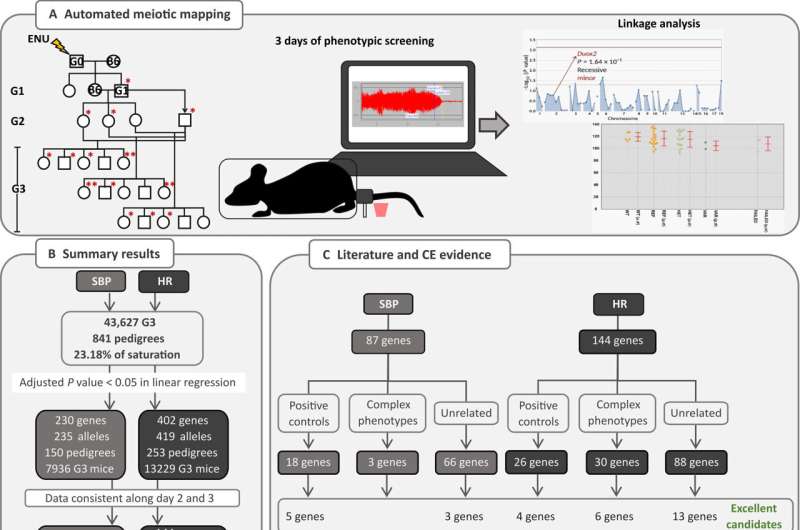
In the recently published study, the researchers combined two techniques to find out which genes are involved in blood pressure and heart rate alterations. First, they used drugs to induce mutations in mice. The mutations were randomly distributed throughout the germline (cells responsible for genetic inheritance). In this stage, it was not yet possible to identify the genes involved in the mutations detected.
Next, the researchers sequenced the exomes of these animals' descendants to find out which genes were linked to the mutations. The exome is the protein-coding portion of the genome and is associated with most disease-related genetic variants.
"At our lab, we combine ordinary mutagenesis—induced by the compound N-ethyl-N-nitrosourea [ENU]—with a technology called automated meiotic mapping [AMM], a computational platform that deploys statistical computing and artificial intelligence to identify instantly which mutation [out of an average of 60 in each family of mice] causes some effect in any process we study. This is how we were able to discover such a large number of novel genes that influence blood pressure and heart rate, in addition to identifying many others that had been described before," Beutler told Agência FAPESP.
The numbers are indeed impressive. To identify the 231 genes linked to blood pressure and heart rate, the researchers sequenced the exomes of generations and generations of mice. They analyzed 878 pedigrees (murine family trees) comprising a total of 45,261 animals and detected 45,000 mutations induced by the mutagenic agent (ENU), which damaged 22% of the murine genome and caused phenotype variations.
According to Krieger, the strategy used in the study was carefully designed to enable the researchers to identify genetic variants that control complex phenotypes such as blood pressure and heart rate.
"The methodology enabled us to assess 30 to 60 mutations in gene clusters simultaneously, resulting in a substantial efficiency gain. This was what made it possible to discover so many novel genes associated with factors as significant as blood pressure and heart rate in one fell swoop. What's more, in the dataset we've analyzed so far, we've addressed only 22% of the genome [i.e. the exome], so even more remains to be discovered," he said.
In addition to the discovery of a large number of genes, he continued, the study reported in Science Advances proves the efficacy of germline mutagenesis as a technique for defining key determinants of polygenic phenotypes. It is worth noting that the same animals with mutations produced by the platform are used in studies of different diseases.
"Because mutations are created randomly in the murine germline, and alterations in a given pathological or physiological process, such as maintenance of blood pressure or heart rate, are then detected, we can discover many—and possibly all—of the genes essential to those processes," Beutler said.
According to Krieger, the next step will be to analyze the role of each gene in depth. "We now have a long road ahead of us to find out how these genes do what they do, and whether they can be targets for novel therapies to treat high blood pressure or cardiac arrhythmia, for example," he said.
"Understanding the mechanism remains the bottleneck. When you discover that damage to a certain gene results in a faster heart rate, for example, it's a daunting challenge to understand why. Once this understanding is achieved, however, it's possible to design novel therapeutic strategies to control heart rate or blood pressure," Beutler said.
More information: Samantha K. Teixeira et al, Genetic determinants of blood pressure and heart rate identified through ENU-induced mutagenesis with automated meiotic mapping, Science Advances (2024). DOI: 10.1126/sciadv.adj9797