This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
New synapse type discovered through spatial proteomics
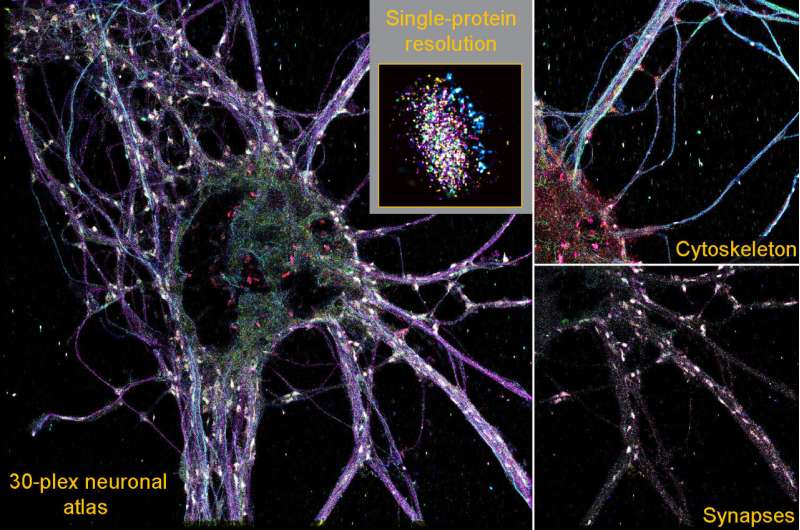
Researchers have developed a new super-resolution high-throughput imaging method. Using the new technique, the scientists were able to create a 3D neuronal cell atlas with single-molecule resolution and discovered a previously unknown type of synapse. The results of the study were published in the journal Cell.
In the current study, led by Eduard Unterauer in Jungmann's laboratory at the MPI of Biochemistry and the LMU, the researchers present SUM-PAINT. This is a new technological development in super-resolution microscopy that, for the first time, allows very fast and virtually unlimited visualization and mapping of a large number of proteins.
"The complexity of living systems ranges from entire organisms and tissues, to the structure of intricate cellular networks, to the organization and interaction of individual biomolecules," says Eduard Unterauer, co-first author of the study.
"To understand this complexity in its entirety, the position, identity, and interaction of individual biomolecules must be studied simultaneously. Such methods, which combine multiple signals, are called multiplexing methods. Four critical challenges must be overcome to achieve a comprehensive understanding of protein organization: Sensitivity, throughput, spatial resolution and multiplexing capability."
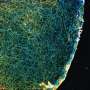
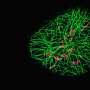
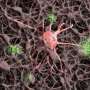
Focusing on the complex environment of neuronal cells in the brain, the team created the first-ever neuronal atlas with single-molecule resolution for 30 different protein types. With improved throughput and multiplexing capabilities, they were able to unravel the complexity of the synaptic protein composition of nearly 900 individual synapses.
To further explore these large data sets, the research team developed a machine learning-based analysis pipeline. By analyzing 1,600 features from the imaging datasets, such as protein content, distribution, or shape, the scientists discovered a previously unknown type of chemical synapse. These synapses make up only about 1% of all synapses. They would not have been detected with other imaging techniques.
With SUM-PAINT, the team provides an integrated workflow for data generation and analysis that can be used by researchers around the world. SUM-PAINT is relatively easy to use with commercially available microscopes.
"We are convinced that SUM-PAINT is not only a milestone on the way to deciphering the complexity of cell biology at the molecular level, but also a potential breakthrough in the discovery of new therapeutic approaches for neurodegenerative diseases," says Ralf Jungmann, head of the Molecular Imaging and Bionanotechnology research group at MPI of Biochemistry and holder of the Chair of Molecular Physics of Life at LMU.
By providing a detailed view of the location and interaction of a large number of proteins at the molecular level, SUM-PAINT opens up unprecedented opportunities to investigate previously hidden details of neurological disorders. In this way, the new method could contribute to a deeper understanding of the underlying mechanisms of diseases such as Parkinson's or Alzheimer's dementia.
More information: Eduard M. Unterauer et al, Spatial proteomics in neurons at single-protein resolution, Cell (2024). DOI: 10.1016/j.cell.2024.02.045