This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
New technique opens up study of immune response to cancer based on DNA locked in old archive cancer samples
Scientists have developed an improved technique for reading the genetic material associated with the body's immune response to cancer, making it possible to study tissue samples that have been stored in archives for decades.
Using this technique, which is much more sensitive than commercial equivalents and less costly, researchers are able to analyze archival cancer samples that would previously have been considered too degraded for analysis.
According to the researchers from The Institute of Cancer Research, London, and University College London who developed the technique, being able to read the vast archives of patient samples available in hospitals around the world will significantly expand opportunities to track how the immune system responds during the development, progression and treatment of cancer. In the long term, this could lead to new ways to detect cancer progression and, ultimately, to more effective treatments.
The research team is also currently seeking a commercial partner to help develop the technique into a widely used tool for lab researchers working in immunology. The study was published in the journal Cancer Research.
Understanding the immune response to cancer
Researchers are continually learning more about how the immune system tries to defend the body against cancer, and the ways in which cancer can evade immune cells or even co-opt them to promote its own growth. The understanding of the interactions between cancer and immune cells, including how they change as cancers develop and respond to treatments, is important for identifying new treatment approaches.
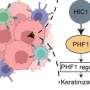
A common area of focus is the T-cell receptor (TCR), a protein complex present on the surface of white blood cells called T cells. TCRs recognize markers on "foreign" cells and cause the immune system to attack them. TCRs can also recognize cancer cells as foreign, thereby playing a key part in the immune system's fight against the disease.
The immune system contains millions of different T cells, each with a unique TCR that recognizes a different marker. Understanding the huge diversity of TCRs is key to determining how the immune system is attempting to defeat the cancer.
Genetic sequencing is used to read the TCRs present in an individual's immune system. However, the existing TCR sequencing methods have a low success rate when used on typical hospital samples, which have been preserved in formalin then paraffin wax. The formalin degrades the genetic material, making it hard to read.
Solving the problem by creating a novel protocol
Recognizing the urgent need for an improved sequencing methodology, the researchers behind the current study developed a new technique called FFPE-suitable Unique Molecular idEntifier-based TCRseq, or FUME-TCRseq.
FUME-TCRseq makes use of a very sensitive lab technique called polymerase chain reaction (PCR) to produce huge numbers of copies of a small section of the TCR that is most important for recognizing foreign cells.
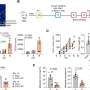
FUME-TCRseq performed well against industry standards
The team was pleased to find that the new protocol outperformed existing commercial methods for TCR sequencing.
The ICR is seeking a collaborative partner for the licensing and co-development of FUME-TCRseq to help make the technique more widely available so that it can benefit cancer research and people with cancer. The institute is particularly interested in out-licensing the technology to a partner who can work with them on the development of FUME-TCRseq.
The ICR is a leader in this field, and its researchers have been involved in many key research advances. These include using AI to access the DNA of FFPE samples, which could allow scientists to track cancer evolution on an unprecedented scale, and developing a way to predict which patients are likely to benefit from immunotherapy—a type of cancer treatment that enhances the body's immune response.
Many possible applications across the health sector
First author Dr. Ann-Marie Baker, Senior Staff Scientist in the Genomics and Evolutionary Dynamics Group at The Institute of Cancer Research, London, said, "FUME-TCRseq is a robust and accurate TCR sequencing methodology able to analyze the TCR repertoire in archival clinical specimens. Using this technique, we can quantify and track specific T-cell populations through both time and space, seeing how they respond to treatment. What is most exciting is that it works really well on archival FFPE material, which a lot of existing methodologies do not."
Senior author Professor Trevor Graham, Professor of Genomics and Evolution and Director of the Center for Evolution and Cancer at the Institute of Cancer Research, London, said, "There's a critical need for a reliable, affordable and accurate method that researchers can use to analyze the low-quality pathology samples that we currently can't access but that may contain important information about how our immune system responds to cancer. We believe these often disregarded archive samples in hospitals around the world could contain a treasure trove of knowledge that will help us in our mission to defeat cancer."
Dr. Iain Foulkes, Director of Research and Innovation at Cancer Research UK, said, "Immunotherapies are becoming more widely available. But they don't work for everyone because, on the surface, cancer cells can look like normal cells. Getting the immune system to recognize and attack cancer is one of the biggest challenges in cancer research today.
"Cracking the complex code of T-cell receptors is vital to improving immunotherapy. This technique opens up a vast archive of samples which could be harnessed to deliver better immunotherapy faster."
More information: Ann-Marie Baker et al, FUME-TCRseq enables sensitive and accurate sequencing of the T-cell receptor from limited input of degraded RNA, Cancer Research (2024). DOI: 10.1158/0008-5472.CAN-23-3340