This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Research team builds first tandem repeat expansions genetic reference maps
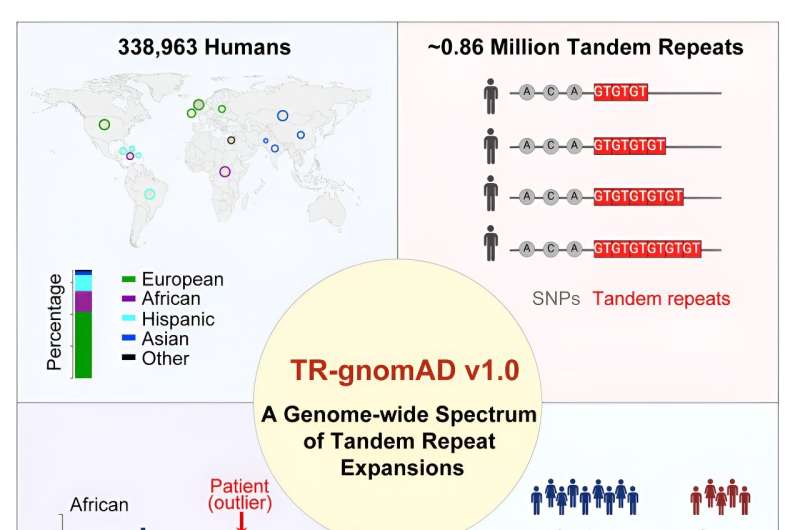
A research team led by the University of California, Irvine has built the first genetic reference maps for short lengths of DNA repeated multiple times which are known to cause more than 50 lethal human diseases, including amyotrophic lateral sclerosis, Huntington's disease and multiple cancers.
The UC Irvine Tandem Genome Aggregation Database enables researchers to study how these mutations—called tandem repeat expansions—are connected to diseases, to better understand health disparities and to improve clinical diagnostics.
The study, published online today in the journal Cell, introduces the UC Irvine TR-gnomAD, which addresses a critical gap in current biobank genome sequencing efforts. Although TR expansions constitute about 6 percent of our genome and substantially contribute to complex congenial conditions, scientific understanding of them remains limited.
"This groundbreaking project positions UC Irvine as a leader in human and medical genetics by addressing the critical gap in the ability to interpret TR expansions in individuals with genetic disorders," said Wei Li, the Grace B. Bell Chair and professor of bioinformatics and co-corresponding author.
"The TR-gnomAD advances our ability to determine how certain diseases might affect diverse groups of people based on variations in these mutations among ancestries. Genetic consulting companies can then develop products to interpret this information and accurately report how certain traits might be linked to different groups of people and diseases."
To build the database, the team utilized two software tools to analyze the genomic data of 338,963 participants across 11 sub-populations. Of the .91 million TRs identified, .86 million were of high enough quality to be retained for further study. It was also discovered that 30.5 percent of them had at least two common alternative forms of a gene caused by a mutation located in the same place on a chromosome.
"Although we've successfully genotyped a substantial number of TRs, that is still just a fraction of the total number in the human genome," Li said. "Our next steps will be to prioritize the integration of a greater number of high-quality TR and include more underrepresented ancestries, such as Australian, Pacific Islander and Mongolian, as we move closer to realizing personalized precision medicine."
More information: Ya Cui et al, A genome-wide spectrum of tandem repeat expansions in 338,963 humans, Cell (2024). DOI: 10.1016/j.cell.2024.03.004