Predicting immune system responses to various stimuli
Just like some people, macrophages—tiny cells that provide the immune system with a primary line of defense against pathogens—reveal a lot about themselves when challenged. Computer scientists and biologists at Pacific Northwest National Laboratory have identified a basic response mechanism that underlies how macrophages respond by presenting the cells with three different insults.
The macrophage immune response is a primary line of defense against infectious disease and inert foreign materials such as environmental particulates. Determining common patterns of activation and important regulatory control points will provide significant avenues for development of therapeutic treatments for a variety of diseases.
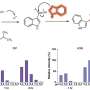
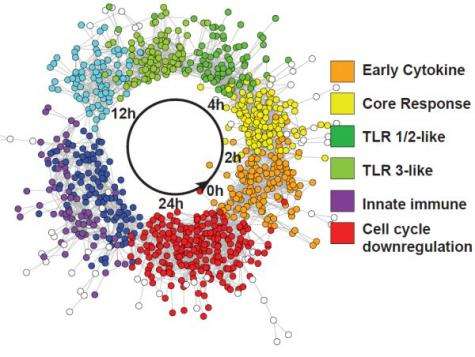
Although macrophages mount different immune responses appropriate to the type of challenge encountered, a core defense response may exist that is activated by many different stimuli. Transcriptomic datasets from macrophages responding to a variety of stimuli, including viral and bacterial components, silica nanoparticles, and infection by Salmonella enterica serovar Typhimuirum, were used to determine similar elements and patterns of defense response networks. These networks were analyzed using a computational network analysis approach and it was found that genes with high topological betweenness in the networks (bottlenecks) are enriched in genes known to be interaction targets of pathogens.
Bottlenecks are highly central to networks and it is believed that they mediate transitions between system states. Next, a comparative analysis method was employed to identify genes responding to multiple stimuli. This analysis revealed a core response module of genes responding to all stimuli examined.
The dynamic behavior of this core response module was modeled using a technique known as multivariate regression, which infers relationships between transcription factors—proteins that bind to specific DNA sequences—and target genes and represents these relationships as ordinary differential equations. The equations then allow prediction of gene expression under conditions not used for training the model. The model was validated by predicting expression of the module responding to nanoparticle exposure. Macrophage responses to nanoparticle exposure are thought to be very different from response to pathogens, and this study provided the first comparison between these two important responses. This is the first report of the application of this approach to data from a vertebrate.
The next steps for these studies are to examine the responses of macrophages to pathogens to distinguish patterns that are caused by actions of the pathogen from those that are common host responses, and to experimentally validate predicted regulatory control points.
More information: McDermott JE, Archuleta M, Thrall BD, Adkins JN, Waters KM (2011) Controlling the response: Predictive modeling of a highly central, pathogen-targeted core response module in macrophage activation. PLoS One 6 (2): e14673.