Discovering new uses for old drugs
With the cost of putting a single new drug on the pharmacy shelves topping a staggering $1 billion, scientists are reporting development of a way to determine if an already-approved drug might be used to treat a different disease. The technique for repurposing existing medicines could cut drug development costs and make new medicine available to patients faster, they report in ACS' Journal of Medicinal Chemistry.
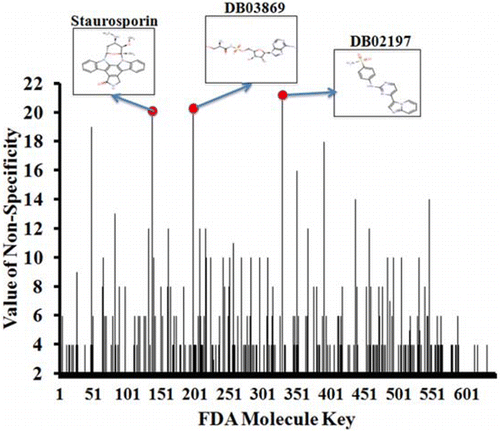
Sivanesan Dakshanamurthy and colleagues explain that drug companies must limit efforts to market new drugs because the current approach is so expensive, time-consuming and prone to failure. Scientists long have known that drugs already approved for one disease might be effective for others. However, existing methods to identify new uses for old drugs lack accuracy and have other disadvantages. So Dakshanamurthy's team developed a comprehensive new computer method called "Train-Match-Fit-Streamline" (TMFS) that uses 11 factors to quickly pair likely drugs and diseases.
They describe using TMFS to discover evidence that Celebrex, the popular prescription medicine for pain and inflammation, has a chemical signature and architecture suggesting that it may work against a difficult-to-treat form of cancer. Likewise, they found that a medicine for hookworm might be repurposed to cut off the blood supply that enables many forms of cancer to grow and spread. "We anticipate that expanding our TMFS method to the more than 27,000 clinically active agents available worldwide across all targets will be most useful in the repositioning of existing drugs for new therapeutic targets," they said.
More information: “Predicting New Indications for Approved Drugs Using a Proteochemometric Method” J. Med. Chem., Article ASAP. DOI: 10.1021/jm300576q
Abstract
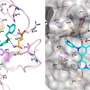
The most effective way to move from target identification to the clinic is to identify already approved drugs with the potential for activating or inhibiting unintended targets (repurposing or repositioning). This is usually achieved by high throughput chemical screening, transcriptome matching, or simple in silico ligand docking. We now describe a novel rapid computational proteochemometric method called “train, match, fit, streamline” (TMFS) to map new drug–target interaction space and predict new uses. The TMFS method combines shape, topology, and chemical signatures, including docking score and functional contact points of the ligand, to predict potential drug–target interactions with remarkable accuracy. Using the TMFS method, we performed extensive molecular fit computations on 3671 FDA approved drugs across 2335 human protein crystal structures. The TMFS method predicts drug–target associations with 91% accuracy for the majority of drugs. Over 58% of the known best ligands for each target were correctly predicted as top ranked, followed by 66%, 76%, 84%, and 91% for agents ranked in the top 10, 20, 30, and 40, respectively, out of all 3671 drugs. Drugs ranked in the top 1–40 that have not been experimentally validated for a particular target now become candidates for repositioning. Furthermore, we used the TMFS method to discover that mebendazole, an antiparasitic with recently discovered and unexpected anticancer properties, has the structural potential to inhibit VEGFR2. We confirmed experimentally that mebendazole inhibits VEGFR2 kinase activity and angiogenesis at doses comparable with its known effects on hookworm. TMFS also predicted, and was confirmed with surface plasmon resonance, that dimethyl celecoxib and the anti-inflammatory agent celecoxib can bind cadherin-11, an adhesion molecule important in rheumatoid arthritis and poor prognosis malignancies for which no targeted therapies exist. We anticipate that expanding our TMFS method to the >27 000 clinically active agents available worldwide across all targets will be most useful in the repositioning of existing drugs for new therapeutic targets.