Improved molecular tools streamline influenza testing and management
Over 40,000 people die each year in the United States from influenza-related diseases. In patients whose immune systems are compromised, antiviral therapy may be life-saving, but it needs to be initiated quickly. It is therefore crucial to diagnose and type the influenza rapidly. Scientists in the Netherlands have designed and evaluated a set of molecular assays that they say are a sensitive and good alternative for conventional diagnostic methods and can produce results in one day without the need for additional equipment. The results are published in The Journal of Molecular Diagnostics.
Currently the main circulating influenza viruses that cause disease in humans are the influenza A H3N2 and H1N1 subtypes together with influenza B virus. Re-emergence of a variant of the H1N1 influenza virus, which circulated in the population between 1977 and 2009, can also not be ruled out. Strategies to combat influenza virus-induced disease rely on vaccination as a preventive measure. In cases where vaccine efficacy is low, antiviral drugs may be used as prophylaxis.
Traditionally the adamantane and neuraminidase inhibitor class of drugs are available for both treatment and prophylaxis. However some subtypes are resistant to these. Most of the recently circulating influenza viruses are resistant to the adamantanes. In addition, the pre-pandemic H1N1 viruses, which emerged at the end of 2007, are naturally resistant to the neuraminidase inhibitor oseltamivir (Tamiflu®). Sensitive and reproducible molecular assays are therefore essential for diagnosing influenza virus subtypes.
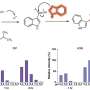
The investigators report on the design, validation, and evaluation of a set of real-time polymerase chain reaction (RT-PCR) assays for quantification and subtyping of human influenza A and B viruses from patient respiratory material, as well as four assays for detecting drug resistant mutations. For the evaluation of these assays, 245 respiratory specimens from 87 patients living in Asia, Europe, and the United States who were enrolled in a prospective study of influenza illness, including assessment of neuraminidase resistance, were analyzed. In addition 96 pre-pandemic influenza A/H1N1 viruses from the epidemic of 2007-2008 were analyzed by the H275Y assay to check the robustness of the assay.
The influenza quantification assay was used to check for virus positivity and to obtain virus particle counts for all analyzed samples. Influenza A viruses were then subtyped and tested for presence of oseltamivir resistance mutations using the resistance RT-PCR assays. In total, 129 respiratory specimens tested positive for influenza A and 60 for influenza B virus. One sample tested positive for both virus types.
"RT-PCR based assays have become the standard in most diagnostic laboratories worldwide in recent years," comments lead investigator Martin Schutten, PhD, Head of the Clinical Virology Unit at the Erasmus University, Rotterdam, the Netherlands. "The assays described here cover all currently circulating human influenza viruses and can detect major resistance mutations to oseltamivir. By introducing external quantification and internal standards, longitudinal assay performance can be monitored carefully and a virus particle count can be assigned to an analyzed sample.
"This algorithm can generate useful data to assist in the management of individual influenza virus infected patients and to evaluate clinical trials. Information regarding influenza virus (sub) type, viral load and antiviral susceptibility can be obtained within one working day. Alongside previously described assays that detect antiviral resistance associated mutations in 2009 pandemic H1N1 virus, these assays are a powerful tool for the clinical management of influenza virus infected patients," he concludes.
Although infection from H7N9, the new potential pandemic Influenza strain, or H5N1, a continuing pandemic threat since 1997, can be identified by exclusion (positive in the Influenza matrix RT-PCR but negative in RT-PCR typing), development of rapid typing RT-PCR for these potential pandemic viruses may be useful in complementing the existing set.
More information: "Molecular Assays for Quantitative and Qualitative Detection of Influenza Virus and Oseltamivir Resistance Mutations: Quantitative Influenza Virus Diagnostics," by Erhard van der Vries, Jeer Anber, Anne Van der Linden, Yingbin Wu, Jolanda Maaskant, Ralph Stadhouders, Ruud van Beek, Guus Rimmelzwaan, Albert Osterhaus, Charles Boucher, and Martin Schutten, dx.doi.org/10.1016/j.jmoldx.2012.11.007. The Journal of Molecular Diagnostics, Volume 15, Issue 3 (May/June 2013)