Quadratic leap in detecting smallest unit of genetic change
Scientists at Rice University and the University of Washington (UW) this week unveiled a groundbreaking new method for detecting minute changes known as single nucleotide polymorphisms (SNPs) in the human genome. The human genome has more than 6 billion base pairs, and one of the revelations of modern genomics is that even the slightest change in the sequence—a single-nucleotide difference—can have profound effects.
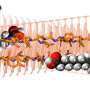
The new SNP genotyping technique, dubbed "double-stranded toehold exchange," is described in a new paper in Nature Chemistry. The patent-pending method is markedly different—in both form and performance—than any of the dozen-plus methods already used to detect SNPs.
"There are two axes of performance in SNP detection—read length and specificity," said study co-author David Zhang, who joined Rice's Department of Bioengineering this month. "We're at least an order of magnitude better on each axis. In fact, in terms of specificity, our theoretical work suggests that we can do quadratically better, meaning that whatever the best level of specificity is with a single-stranded method, our best will be that number squared."
Scientists have sequenced the genomes of dozens of species, but those species-level genomes only tell part of the genetic story for a given individual. In people, for example, slight differences in just a few nucleotides can mean the difference between having green or brown eyes. This type of genetic variation within a species is called polymorphism, and SNPs are the smallest unit of polymorphic variation.
SNPs are the most frequently occurring genetic variation in the human genome; more than 30 million have been confirmed. But they also occur in other species, even in single-celled organisms. In the bacteria that cause tuberculosis (TB), for example, an SNP in the right location can allow the disease to fight off antibiotics like rifampicin, one of the most commonly prescribed anti-TB drugs. Though small on a molecular scale, that single-nucleotide difference has serious implications for TB patients. Rather than going through a six-month course of antibiotics costing about $20, patients with drug-resistant TB often face more than two years of treatment with drugs that sometimes have permanent side effects and can cost more than $2,500.
"Drug-resistant TB is an enormous global problem, and it's growing," said UW synthetic biologist Georg Seelig, the lead researcher on the project. "One reason we chose to test our method on finding SNPs in TB is because those tests are vitally important for the fight against TB."
Existing SNP detection methods have a relatively short read length; this means that scientists might have to run a half-dozen or more of the tests to look for an SNP in a 200-base-pair region of a genome. The Rice-UW method uses a novel approach to allow for much longer read lengths.
"In these tests, our read length was 198 base pairs because that was the length of the region we needed to scan for SNPs related to rifampicin resistance," said Seelig, assistant professor of computer science and engineering and of electrical engineering at UW. "We could have designed a longer probe if we'd needed one. There is no inherent limitation to the length of the probe we can make. Reaction time—not read length—is likely to be the limiter with this method."
Selectivity is also an issue in SNP detection. The more selective a test is, the more likely it is to detect a rarely occurring SNP. For example, a TB patient might be infected with both drug-resistant and non-drug-resistant strains of TB at the same time.
"Maybe only 1 percent of the TB in the patient is resistant to rifampicin," said Zhang, Rice's Ted Law Jr. Assistant Professor of Bioengineering. "If you treat that person with rifampicin, the result is that you will kill the 99 percent and give the drug-resistant variety a chance to become well-established."
An SNP test would need a selectivity of 100-to-1 to accurately diagnose the patient in the above example. Zhang said some current methods can approach that level of selectivity, but only if samples are prepared with painstaking attention to temperature, pH and other conditions.
"Our selectivity was about 12,000-to-1 in this study, and we don't require any special conditions," he said.
Zhang and Seelig said the selectivity of the technique derives from the use of double-stranded probes. Crafting these wasn't easy, they said. The team had to add single-stranded "toeholds" to each end of the double-stranded probe to improve reaction kinetics and to speed the binding process enough to make the test practical.
"We are moving forward with the goal of applying this technology to infectious disease diagnostics," Seelig said.
More information: Nature Chemistry (2013) doi:10.1038/nchem.1713