December 5, 2013 feature
Getting to know you: Directed evolution allows pathobiology-free antibody determination
(Medical Xpress)—Precise detection of specific antibodies is fundamental in diagnosing a wide range of diseases – and testing for antibodies using known antigens is used extensively to diagnose infectious and autoimmune diseases. That being said, developing reagents that test of such antibodies – and antibody identification itself – have historically been a very difficult undertaking. Recently, however, scientists at University of California, Santa Barbara have developed the antibody diagnostics via evolution of peptides (ADEPt) method for simultaneously discovering both antibody disease biomarkers and reagents that accurately detect them. In addition, ADEPt may provides a means of potentially developing effective diagnostic tests where none exist, as well as for identifying environmental disease-related factors.
Prof. Patrick S. Daugherty and Medical Xpress discussed the paper he, researcher John T. Ballew and their co-authors at University of California, San Diego, Mayo Clinic, and University of Tampere, Tampere University Hospital and Seinäjoki Central Hospital, Finland recently published in Proceedings of the National Academy of Sciences. Daugherty first summarized the challenges they faced in their research. Most importantly, Daugherty tells Medical Xpress, it has been very difficult to identify which of the many antibodies in a patient sample is associated with disease. "The main challenge has been to identify the particular antibody types that occur in most or all patients, and to determine which antigens these antibodies recognize. While we have several tools to determine the human autoantigens to which these antibodies bind, the myriad of possible antigens makes finding environmental factors or antigens very difficult." (Autoantigens are antigens that despite being normal tissue are the target of an immune response that gives rise to autoimmune disease.) "On top of that," he adds, "even if one antigen gives rise to the disease antibodies, each person can make a slightly different version of antibody to that antigen." In other words, the antibody in one patient will not be identical to the antibody in another.
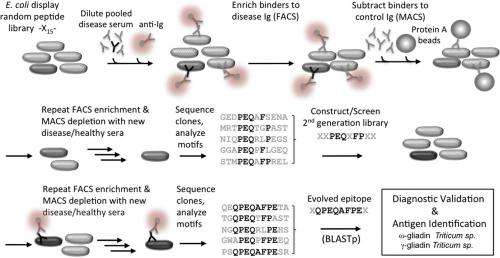
"This is really a molecular needle in a haystack type problem," Daugherty continues. "In order fish out the disease antibodies and their antigen partners, we had to develop a more sophisticated molecular separation process." Their approach was to use E. coli bacteria that display billions of possible peptide antigens on their outer surface – a collection of candidates called a library. They then select those cells that bind to the patient's antibodies with a cell sorter. "However," Daugherty points out, "the problem isn't so simple: We might be left with millions of peptide-antibody pairs, most of which have nothing to do with the disease in question." To address this, the scientists removed binders to antibodies from healthy controls, and repeated these steps with five additional disease and control samples. The final result was that the researchers had performed roughly 15 individual separation steps to zero in on the peptide-antibody pairs that occur only in the setting of celiac disease.
"Our second realization was that whatever peptides we retrieved from our peptide libraries, they were not going to be optimal." Daugherty says. "They're more like draft solutions that attempt to conform to the precise shape and chemistry of the antibody binding pocket." As an example, he illustrates, the rarest string of even eight amino acids in their peptide library will occur at a frequency of about one in a trillion molecules – but since they can only can make about 10 billion, they can reasonably conclude that the best solution will not be in the original library.
To tackle this problem, the scientists turned to a widely employed biotechnology method known as directed in vitro molecular evolution, which mimics natural evolution in the laboratory but operates on a molecular level. The researchers used information from the first generation peptides to design a more focused library, which was then subjected to the molecular separation process. "In this way we could find those one-in-a-trillion peptide sequences even though our libraries were only numbered about a billion – and what we found excited us," Daugherty tells Medical Xpress. "The peptides from this evolution process converged upon a particular 8-amino acid string, and one of these evolved peptides was able to detect and capture disease antibodies from all of our subjects with celiac disease but did not react with our controls." In short, molecular evolution found very rare peptides to which all patients developed an immune response, meaning that the researchers had effectively built a diagnostic reagent from scratch that could detect the celiac disease-specific antibodies present in nearly all of the subjects used in their study.
"It's very difficult to discover and develop reagents that can detect all patients' antibodies directed towards the same antigen," Daugherty explains. "If many different individuals are exposed to one protein – for example, from a vaccine antigen – particular parts of those proteins called epitopes might be targeted by their immune systems – but because there are so many different ways to make an antibody, each person's antibody is slightly different, even in response to the same epitope. To create a single diagnostic test that detects all patients' antibodies it is necessary to discover or create a detection reagent (in this case a peptide) that closely mimics the unidentified antigen to which each person is responding – and if the shared antigen were it identified, it would be expected to provide the highest accuracy." The problem is that in most cases the antigen that is the target of the disease-specific antibodies is unknown. The method presented in this study, however, effectively creates a draft molecular mimic of the true antigen and then iteratively optimizes this draft. The result is a single optimized peptide that detects a maximum number of patients' antibodies.
"This iterative optimization process, when applied to blood antibodies from celiac patients, dramatically increased the diagnostic accuracy of the reagents and simultaneously converged on a sequence that matched the environmental antigen – namely, deamidated gliadins (a modified form of wheat gliadin protein)" Daugherty continues, "Moreover, our method could be applied to just about any human tissue fluids that contain antibodies. Aside from using serum or plasma, it would be interesting to look at antibodies present in saliva, some of these antibodies may be similarly diagnostic useful but would not require any blood collection. Another option would be look at extracts from tissues where the disease process is active." For example, he notes that in rheumatoid arthritis analysis of synovial fluid present in inflamed joints might provide addition clues that are not apparent from blood analysis, and adds that analysis of other human tissue fluids could also be useful – such as skin interstitial fluid, which might reveal antibodies produced in response to skin irritations or infections.
"Initially, our primary focus was to develop a capability to create low cost diagnostic tests – but we've stumbled across something that may have greater significance," Daugherty relates. "When we searched our evolved peptides against the entire protein database that contains all of the proteins known from tens of thousands of different organisms whose genome has been sequenced, the very best matches were from proteins in wheat, barley and rye – the known environmental factors in celiac disease." While there are available diagnostic tests that detect these antibodies, they've been developed only through many years of painstaking R&D – but the researchers arrived at the same answer through a completely different process that, remarkably, is blind to which antibodies or antigens might be involved in celiac disease.
They also found a second type of peptide that only celiac disease patients will respond to and produce antibodies. "We suspect that this peptide reflects another environmental factor – and, possibly, a microorganism that lives in the gut. If we can identify the antigen, it might provide clues that lead to a better understanding of celiac disease, or development of celiac disease therapeutics." Daugherty adds that they're now trying to evolve this peptide sequence in an effort to match it to an environmental factor.
"Our excitement about this method is that we may be able to find previously unknown environmental factors for many other diseases," says Daugherty. The scientists are have already seen early benefits from applying their process to blood specimens from women with the common but serious pregnancy condition known as preeclampsia. They're also building alliances with clinicians to search for environmental factors involved in other autoimmune diseases as well as neurological and psychiatric diseases that may have an autoimmune component.
"Moving forward, we really want to build a more comprehensive picture of the immune response, and specifically the antibody repertoire," Daugherty continues. "To do this, we hope to catalog thousands of different types of antibodies that occur in each person, and eventually match each antibody back to an antigen. Longer term, we may be able to map each person's set of antibodies back to their diverse environmental exposures, or in effect, decode immunological memory."
Longer term, Daugherty foresees a handheld device that could test for thousands of different environmental exposures with a drop of blood, enabling diagnosis of hundreds of diseases, food allergies, drug sensitivities, and prior vaccinations." "Once the peptide sequences that each antibody species binds to are known, they could be incorporated into a single diagnostic device that could look for the presence of thousands of environmental factors to which an individual has responded."
Daugherty also sees many areas of medicine that might benefit from their technology. "We can already see the possibility for new therapeutic targets and strategies arising from the discovery of the antibody-antigen pairs" he points out. "If we identify and validate environmental triggers, this could open up the possibility for new therapies or vaccines that might block these triggers. I think that this type of antibody repertoire analysis could be used to design vaccines or characterize vaccine efficacy and safety in greater detail. One could obtain precise functional signatures of individual responses to vaccines in the form of antibody-peptide antigen pairs," he concludes, "and these signatures could be compared between individuals and correlated with their clinical response."
More information: Antibody biomarker discovery through in vitro directed evolution of consensus recognition epitopes, PNAS Published online before print November 12, 2013, doi:10.1073/pnas.1314792110
© 2013 Medical Xpress. All rights reserved.