New atlas of molecules paves the way for improved understanding of metabolic diseases
In the most comprehensive exploration of the association between genetic variation and human metabolism, researchers have provided unprecedented insights into how genetic variants influence complex disease and drug response through metabolic pathways.
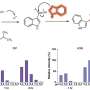
The team has linked 145 genetic regions with more than 400 molecules involved in human metabolism in human blood. This atlas of genetic associations with metabolism provides many new opportunities to understand the molecular pathways underlying associations with common, complex diseases.
Metabolic molecules, known as metabolites, include a wide range of different molecules such as vitamins, lipids, carbohydrates and nucleotides. They make up parts of, or are the products of, all biological pathways. This new compendium of associations between genetic regions and metabolite levels provides a powerful tool to identify genes that could be used in drug and diagnostic tests for a wide range of metabolic disorders.
"The sheer wealth of biological information we have uncovered is extraordinary," says Dr Nicole Soranzo, senior author from the Wellcome Trust Sanger Institute. "It's exciting to think that researchers can now take this freely available information forward to better understand the molecular underpinnings of a vast range of metabolic associations."
The team measured the levels of a large number of metabolites, both those already known and many as yet uncharacterised, from many different metabolic pathways.
They found 90 new genetic associations, trebling the figure of known genetic associations with metabolites. In many of the cases where metabolites were known, the team were able to link the molecule to gene function. They mapped genes to their likely substrates or products and linked these to a number of conditions, including hypertension, cardiovascular disease and diabetes.
They further found that these genetic regions map preferentially to genes that are currently targeted in drug-development programmes. This provides new opportunities to assess genetic influences on drug response, and to assess the potential for existing drugs to treat a wide range of diseases.
"We developed an open-access database that allows researchers to easily search through the findings, to understand genetic variants associated with metabolism one metabolite at a time and in the context of the complete metabolic network," says Dr Gabi Kastenmüller, co-senior author from the Helmholtz Center Munich, Germany. "This database will facilitate drug discovery for metabolic disorders and also help researchers to understand the biology behind disease."
Other associations suggest tantalising possibilities for further study. For instance, a number of the genetic associations identified involved aromatic acids, such as tryptophan, which are important for brain function. While this study did not measure association of metabolites in the brain, these genetic findings open new avenues to assess potential genetic influences on brain function and responses to drugs that affect brain function, such as antidepressants.
"This work provides an important new window into the genetic variation underlying human metabolism," said Dr Eric Fauman, study co-author and Associate Research Fellow from Pfizer Inc. "Through targeted Precision Medicine and by linking human disease genes to in vivo biological markers, we hope to enhance our ability to deliver impactful new medicines for patients across a variety of disorders."
More information: Shin S-Y et al (2014) An atlas of genetic influences on human blood metabolites. Nature Genetics. DOI: 10.1038/ng.2982