Large-scale study raises hopes for development of E. coli vaccine
The largest ever study of the bacterium enterotoxigenic Escherichia coli (ETEC) has raised hopes that a global vaccine can be developed. This bacterium causes 400 thousand deaths and 400 million cases of diarrhoea each year in low- and middle-income countries as well as misery to many travellers to these regions.
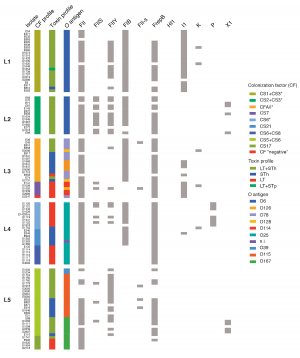
Analysis based on whole-genome sequence data for 362 different ETEC strains that span 30 years and 20 countries has overturned previous thinking about the bacterium's diversity. By comparing the way strains attach to human stomach lining, driven in part by the colonisation factor, researchers found that strains clustered into surprisingly closely related groups. This means that the bacteria that cause disease in countries in Latin America are actually very similar to those in Africa and Asia. Some E. coli have spread globally from a common source, a previously unrecognised phenomenon.
"This research strengthens our belief that it is possible to target a broad range of ETEC groups with one vaccine," says Professor Gordon Dougan, senior author from the Wellcome Trust Sanger Institute. "By targeting the most prevalent colonisation factors in these lineages, we stand a chance of developing a vaccine that will reduce the disease burden caused by this bacterium. This work is now underway at the Sanger Institute."
The close relationship between samples in Asia, Africa and the Americas suggests that a vaccine could be effective worldwide. Samples taken from children and adults in regions where the diarrhoeal disease is endemic and from people who had travelled to these areas were also similar, suggesting that a potential vaccine would work across patient groups.
Large-scale studies like this enable researchers to plot the phylogenetic structure, the pattern of global emergence and evolution, of lethal bacteria such as ETEC. While ETEC was previously thought to vary widely across the world, this phylogeny traces many of the 21 different lineages back their origins in an individual bacterium that acquired the genetic information required to infect humans and then spread, in part due to global travel. According to this data, the emergence of these lineages probably took place between 51 and 170 years ago, a finding that suggests that the lineages are stable and unlikely to become quickly resistant to vaccines.
"Our work to develop a vaccine has received a real boost from this research," says Dr Åsa Sjöling, an author from the University of Gothenburg and Karolinska Institutet, Sweden. "This data gives us confidence that a global, future-proof solution is within our grasp."
Detailed genetic information on each lineage has been made publicly available to the research community, enabling scientists to learn more about the bacteria and to track its future spread and evolution. Among other studies, researchers will use this data to learn more about the 130 ETEC samples that had colonisation factors that had never been identified before.
"This dataset is exceptional," says Astrid von Mentzer, first author from the University of Gothenburg and the Sanger Institute. "Researchers around the world will be able, from these whole-genome sequences, to draw out data about the virulence profiles and other proteins that may be important for virulence and that these strains could have in common. All of this research helps us to identify the shared weaknesses in ETEC bacteria that we can exploit with vaccines."
"Whole genome sequencing has enabled us to quantify what makes an ETEC in unprecedented detail; information that is vital in order to target the development of new vaccines," says study co-author Dr Thomas Connor from Cardiff University. "The challenge now is to distil, from the vast wealth of data locked up in this dataset, the information needed to develop these new treatments to fight this key pathogen."
More information: "Identification of enterotoxigenic Escherichia coli (ETEC) clades with long-term global distribution." Nature Genetics (2014) DOI: 10.1038/ng.3145