Studies of mutated protein in Lou Gehrig's disease reveal new paths for drug discovery
Several genes have been linked to ALS, with one of the most recent called FUS. Two new studies in PLoS Biology, one from the University of Pennsylvania School of Medicine, and the other from colleagues at Brandeis University, both examined FUS biology in yeast and found that defects in RNA biology may be central to how FUS contributes to ALS, or Lou Gehrig's disease. These findings point to new targets for developing drugs.
Proteins aggregate to form insoluble clumps in the brain and spinal cord of ALS patients. In some instances of ALS, the clumping protein is FUS, while in other cases it is another protein called TDP-43. FUS and TDP-43 are both RNA-binding proteins with similar features. For example, both proteins contain a region that is remarkably similar to the type of section that enables some proteins to form prions in yeast. Prions are rogue infectious proteins that cause mad cow disease in cattle and Creutzfeldt–Jakob disease in humans. Despite these similarities it was not clear if TDP-43 and FUS both contribute to ALS in similar or different ways.
In 2009, two groups found mutations in the FUS gene in some ALS patients. In the same year, co-senior author Aaron Gitler, PhD, assistant professor of Cell and Developmental Biology, used a yeast model to study FUS and to determine what effect those mutations were having on its function. Meanwhile, co-senior author James Shorter, PhD, assistant professor of Biochemistry and Biophysics, purified the FUS protein and studied the properties that made it readily form clumps. Gitler and Shorter had previously teamed up to study TDP-43 in yeast cells and pure protein assays. "This is an exciting time. The picture is really coming together for the molecular players in ALS" says Gitler.
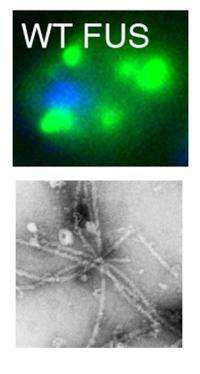
When the researchers overexpressed human FUS in yeast, clumps formed in the cytoplasm of the yeast cells. TDP-43 formed clumps in the yeast cells too. "However, there were important differences in the way in which FUS and TDP-43 clumped in yeast cells, and pure protein assays," explained Gitler lab postdoctoral fellow and co-first author Zhihui Sun, PhD.
FUS is typically found in the nucleus of human cells. In some ALS-associated mutations of FUS, the protein is more abundant in the cytoplasm, suggesting that it is this misplacement that may be causing disease. In line with that, both groups found that restricting overexpressed normal FUS protein to the nucleus decreased clumping. Both groups showed that the biochemical features that promote clumping differ between FUS and TDP-43.
"The prion-like portion is very important for both proteins to clump, but surprisingly and in contrast to TDP-43, FUS needed additional sequences in a separate part of the protein to initiate clumping," says Shorter. "This suggests that the disease-causing mechanism probably differs between ALS associated with FUS clumping versus TDP-43 clumping," explained Shorter lab research specialist and co-first author Zamia Diaz.
"To be honest, we expected the same behavior for TDP-43 and FUS in yeast and at the pure protein level in terms of how the protein clumps. So, next we asked what proteins reverse the toxicity? Do TDP-43 reversers affect FUS clumping toxicity?" says Gitler.
"We were stunned that they were not the same," notes Shorter.
In the FUS screen they found that genes related to cellular structures called stress granules could rescue FUS-related toxicity in yeast cells. These granules sequester RNAs during times of stress for the cell – such as increases in temperature, exposure to toxins, or injury – to use later after stress passes, since cells need to reserve energy. The RNAs are used in the translation of the genetic code into proteins, which uses much of the cell's energy. Mutations in FUS seem to promote it being sequestered in stress granules, and FUS toxicity is associated with these granules. Whether or not these structures played a direct role in FUS toxicity remained unclear. But the new research from both groups suggests that they do play a key role in FUS toxicity. They used genome-wide screens to find genes that could reverse FUS toxicity.
"We were surprised in the genome-wide screens by the lack of overlap in genes that modified FUS toxicity versus TDP-43 toxicity in yeast cells," says Shorter.
"The implication is that we need to target different pathways, that is proteins in FUS-related ALS versus TDP-43–related ALS," adds Diaz.
These studies point the way to new therapeutics for some ALS cases, based on RNA processing. They also suggest caution in the assumption that TDP-43 and FUS both contribute to ALS in the same way. Testing ideas about pathogenesis and treatment is faster and cheaper in yeast, which leads to more rapid progress in understanding the disease and, eventually, its treatment.
"It's truly amazing what we can learn from yeast," concludes Gitler.