Weill Institute researchers uncover basic cell pathway
Although all cells in an organism have the same DNA, cells function differently based on the genes they express. While most studies of gene expression focus on activities in the cell's nucleus, a new Cornell study finds that processes outside the nucleus -- along the cell membrane -- also play important roles in gene expression.
The study, published in the May 1 issue of Genes and Development, uses yeast cells, a simple model system with 6,000 genes, most of which are found in other organisms, including humans, making them excellent candidates for studying complex biological pathways. This paper focuses on pathways related to the Gal1 gene, which is highly researched for characterizing as a model to understand how gene expression is induced and repressed.
The Cornell researchers identified two proteins, Tup1 and Cti6, which form a complex that regulates transcription of the Gal1 gene, but only through interactions with a lipid found on the cell membrane. Tup1 is highly conserved, meaning it has been unchanged through evolution and is found in many organisms from yeast to humans.
"People just focus on the chromosomes inside the nucleus [when studying gene expression]," said lead author Bong-Kwan Han, a research associate at Cornell's Weill Institute for Cell and Molecular Biology, who co-authored the paper with Scott Emr, director of the Weill Institute. "We show that we also have to look at the cytoplasm," Han added.
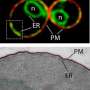
The researchers propose that Cti6 and a complex of the proteins Cyc8 and Tup1 shuttle out of the cell's nucleus to the cell membrane, where they bind to a lipid called PI(3,5)P2 and then further assemble into a Cti6-Cyc8-Tup1 complex. This complex then shuttles back into the nucleus where it binds to other protein structures and plays central roles in activating and repressing transcription, and activating a repressed GAL1 gene.
"Our findings may provide important insights to understand how human Tup1 proteins regulate gene expression in our body," said Han. A few years ago, other researchers reported that the Charcot-Marie-Tooth disorder, a neurodegenerative human disease, may be related to a gene that regulates levels of the PI(3,5)P2 lipid. Also, since human Tup1 proteins are known to play an important role in nerve development, the researchers wonder whether the relationship between the PI(3,5)P2 lipid and Tup1 discovered in this study may be a factor in the underlying mechanism in the Charcot-Marie-Tooth disorder.
Future studies may explore whether such lipids modulate transcriptional regulators in other contexts and whether there are more examples of such lipid-mediated signaling mechanisms that shuttle between the cytoplasm to regulate the nucleus function of transcriptional factors.