Penn Study Explains Paradox of Insulin Resistance Genetics
(Medical Xpress) -- Obesity and insulin resistance are almost inevitably associated with increases in lipid accumulation in the liver, a serious disease that can deteriorate to hepatitis and liver failure. A real paradox in understanding insulin resistance is figuring out why insulin-resistant livers make more fat. Insulin resistance occurs when the body does a poor job of lowering blood sugars.
The signals to make lipid after a meal come from hormones - most notably insulin - and the direct effect of nutrients on the liver. In a recent issue of Cell Metabolism, Morris Birnbaum, MD, PhD, professor of Medicine at the Perelman School of Medicine, University of Pennsylvania, describes the pathway that insulin uses to change the levels of gene expression that control lipid metabolism. Birnbaum is also associate director of the Institute of Diabetes, Obesity, and Metabolism at Penn.
Since insulin normally stimulates fat synthesis in the liver, the expectation is that an insulin-resistant liver would not be able to make lipid. Insulin normally shuts off glucose output and during insulin resistance output is too high. This contributes to the high blood sugar of diabetes. In order to treat the lipid accumulation as well as the glucose abnormalities in type 2 diabetes, it is important to understand the pathways that regulate lipid metabolism.
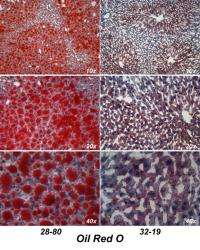
Researchers have suggested that two transcription factors, proteins called FoxA2 and FoxO1, act downstream of, and are negatively regulated by, an enzyme stimulated by insulin called kinase Akt/PKB. Birnbaum had previously shown that this kinase is required for lipid accumulation in the liver. This system is proposed by researchers as a key determinant of liver triglyceride content, one indicator of increased lipids.
In the current study, the team used a technique of introducing mutations into specific genes to show that having these transcriptions factors turned on all the time cannot account for the protection from lipid accumulation in the liver afforded by deleting Akt2 in the liver.
The researchers showed that the major downstream path that insulin uses to regulate these genes converges with the pathways that the body uses to metabolize nutrients. In addition, another arm of insulin signaling (which is probably independent of the nutrient pathway) is also required for the increase in lipid metabolism. Another downstream target turned on by Akt, the mTORC1 protein complex, is required for the body to make lipid. Having multiple pathways is probably a way that the liver makes sure that lipid synthesis is activated only when there is an increase in nutrients and there is a signal from insulin, surmise the researchers.
"Since a therapeutic goal is to prevent this lipid accumulation, any time we identify a novel pathway it raises the hope that there is a previously unknown target out there for a new type of drug," concludes Birnbaum.