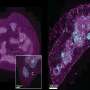
Scientists solve 1,000 protein structures from infectious disease organisms
Investigators at the Center for Structural Genomics of Infectious Diseases (CSGID) and the Seattle Structural Genomics Center for Infectious Disease (SSGCID) announced today that they reached a significant milestone by determining 1,000 protein structures from infectious disease organisms. The knowledge gained from these structures should lead to new interventions for the deadly diseases caused by these pathogens.
Since 2007, the SSGCID, headed by Dr. Peter J. Myler of Seattle Biomedical Research Institute (Seattle BioMed), and the CSGID, headed Dr. Wayne Anderson, Molecular Pharmacology and Biological Chemistry at the Northwestern University Feinberg School of Medicine, have been funded by five-year contracts from the National Institute of Allergy and Infectious Diseases (NIAID), which is part of the National Institutes of Health (NIH).
Researchers in both centers use X-ray crystallography and nuclear magnetic resonance to examine the atomic details of proteins from more than 40 human pathogens, including those responsible for the plague, anthrax, salmonellosis, cholera, tuberculosis (TB), leprosy, amoebic dysentery and influenza. The proteins are selected for their biomedical relevance, as well as potential therapeutic and diagnostic benefits, with one-third being direct requests from the infectious disease research community. "We are laying the groundwork for drug discovery," Anderson says. "Determining protein structures can help researchers find potential targets for new drugs, essential enzymes, and possible vaccine candidates." Myler adds, "The importance of this work is highlighted by the 80+ scientific articles published by the two centers, which also showcase new methodologies developed by each center."
One of the major challenges in medicine today is fighting bacteria that have become drug-resistant. Methicillin-resistant Staphylococcus aureus, commonly known as MRSA, is incredibly difficult to treat because it has developed a resistance to antibiotics, including penicillin and cephalosporins. "By determining the structure of proteins targeted by these drugs, we can now look at how the atoms are arranged in space and how they interact with one another," Anderson says. "Then researchers can determine how the bacterium developed resistance and figure out what to change in the drug so that the bacteria will not recognize it." Dr. Lance Stewart, co-PI of the SSGCID, continues, "The emergence of multi-drug resistant strains of Mycobacterium tuberculosis (MDR-TB), is also an important global health concern, with the most recent cases of TB emerging from India being considered extensively drug resistant (XDR-TB)." The World Health Organization (WHO) and other global health authorities have called for a concerted effort to identify new therapeutic agents for new and better drugs to combat TB, especially medications directed at treating drug resistant strains of the disease. The SSGCID has solved 22 structures from M. tuberculosis and an additional 126 closely related targets from other Mycobacterium species, which cause diseases such as leprosy, Buruli ulcer, and lung infections in AIDS patients. These structures will aid in the understanding of these deadly diseases, as well as providing a blueprint for development of new drugs.
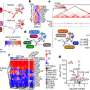
When the two structural genomics centers originally received NIH funding, their expectation was to solve 750 structures, combined, over five years. However, both centers have been driven to exceed this goal by the need for structural data for these pathogens. Advances in technology and the efficiency of both teams allowed them to far exceed their initial goal, even before completion of the 5-year contract. "It used to take four years to determine one structure," Anderson says, "now we can do about three per week." Myler says, "The interaction of our centers with more than two hundred scientific collaborators will greatly accelerate drug development and understanding of the biology of these organisms." Each institution in the two consortia contributes to different aspects of the high-throughput pipeline, which include selecting target proteins using bioinformatics, cloning their genes into bacteria for protein production, purification, and crystallization, with X-ray diffraction data collected at 9 different beam-lines around the United States and Canada. After the protein structures are solved, their coordinates are made available freely to the research community by deposition in the NIH-supported Protein Data Bank. The structures can also be accessed at the CSGID and SSGCID websites.
Members of the scientific community interested in nominating protein targets for structural determination can complete a request form on the SSGCID or CSGID website. This service is offered free of charge to the scientific community, and both centers also freely distribute the protein expression clones via the NIH-funded Biodefense and Emerging Infections Research Resources Repository.