TB outbreaks could be 'solved' by DNA tracking
Reconstructing the spread of killer diseases such as tuberculosis (TB) from person to person using DNA sequencing quickly identifies the origin and movement of pathogens. This approach is directly informing public health strategies to control infectious disease outbreaks, says a scientist speaking at the Society for General Microbiology's Autumn Conference at the University of Warwick.
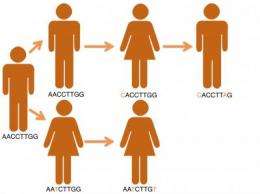
A team from the British Columbia Centre for Disease Control in Vancouver, Canada used whole-genome sequencing to analyse the bacterial DNA in samples from 36 of 41 infected individuals in a TB outbreak. They were able to track the pathogen's movements through the community in British Columbia, including where it started and who infected whom. From this they could identify key persons, places, and behaviours that contributed to the spread of disease. They showed how the social structure of a community contributed to the rapid spread of TB and that a rise in crack cocaine use in the area may have triggered the outbreak.
Earlier epidemiological tools analysed some - but not all - of the DNA in infected samples. This gave too little information to accurately reconstruct an outbreak and scientists could only make informed guesses at how a pathogen spread through a population. Lead researcher Dr Jennifer Gardy, speaking at the conference said, "'Solving' an outbreak - identifying the source of the disease and the underlying patterns of transmission - is the proverbial holy grail of epidemiology. Using whole-genome sequencing we are now able to act like field naturalists and observe how pathogens behave out in the wild. We can see where outbreaks start and how they spread. This level of insight has never been seen before and it promises to change the way we do public heath outbreak investigations."
The group is currently applying the same reconstruction technique to a different TB outbreak in a different social setting. "We are discovering how important location-based transmission is, and that identifying and screening individuals who visited these locations are integral to outbreak management," explained Dr Gardy. Other research groups are applying this genome-based approach for outbreak reconstruction to other micro-organisms. "We hope to build a 'pathogen knowledge base' that describes how different communicable diseases spread in different social and environmental settings. From this we will be able to identify commonalities that can be targeted in global public health interventions."
The work is directly informing public health policy and practice, said Dr Gardy. "As frontline public health practitioners we are using whole-genome sequencing to answer pressing questions. We are able to take our research findings and translate them into clinical action in the short term. With better control and prevention programmes, we can ultimately reduce the burden of certain infectious diseases," she said.
More information: Dr Gardy's presentation "A, C. G, TB: using next-generation sequencing to understand outbreaks and epidemics of tuberculosis" will take place on Tuesday 4 September in session WA05 at the Society for General Microbiology's Autumn Conference 2012.