Non-coding antisense RNA can be used to stimulate protein production
While studying Parkinson's disease, an international research group made a discovery which can improve industrial protein synthesis for therapeutic use. They managed to understand a novel function of non-protein coding RNA: the protein synthesis activity of coding genes can be enhanced by the activity of the non-coding one called "antisense."
To synthesize proteins, the DNA needs RNA molecules serving as short "transcriptions" of the genetic information. The set of all these RNA molecules is called "transcriptome." In the human transcriptome, along with around 25 thousand sequences of coding RNA (i.e. the sequences involved in the synthesis process), an even larger number of non-coding RNA sequences can be found. Some of these RNAs are called "antisense" because they are complementary to sequences of coding RNA called "sense" (the pairing of a sense and an antisense RNA can be seen as a zip).
The RIKEN Omics Science Center has previously discovered that many of the protein coding genes have corresponding antisense RNAs. A study published in Nature, coordinated by a group of SISSA researchers in Trieste, Italy, has now found that a particular type of antisense RNAs stimulate the translation of the protein coding mRNAs that they overlap to. This is in sharp contrast with the current belief that antisense RNAs are universally associated to negative regulation of protein translation.
Most of the mammalian genome is transcribed producing non-coding RNA. The RIKEN FANTOM projects have earlier demonstrated that the largest output of the genome is constituted by non-coding RNAs. More than 70% of the mRNAs are associated in cells with non-coding antisense RNAs, which are usually thought to negatively repress transcription or translation.
In an exceptional collaborative study based on RIKEN FANTOM sense-antisense cDNA clones, the consortium (including SISSA and the RIKEN Omics Science Center) has found a class of non-coding antisense RNAs that do the contrary of what is currently known: enhance translation of mRNAs with which they are pairing. The researchers identified this function studying the antisense of the mRNA of Uchl1, a mouse gene involved in brain function and neurodegenerative diseases. The team, using bioinformatics and data-mining at RIKEN, has also discovered that the antisense of Uchl1 RNA is not a single case but instead is the representative of a larger class of mammalian antisense RNAs, which function is to increase translation. This is the first report of an antisense RNA that increases protein production, which works both in mouse and human cells and is predicted to have similar function in other organisms.
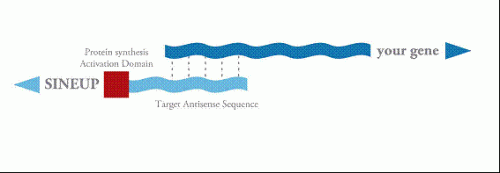
The mechanism to stimulate translation is based on increased association of mRNAs with ribosomes, which is mediated by a SINEB2 element, a repetitive sequence in the antisense of Uchl1 RNA, which is placed in an inverted orientation in the non-coding RNA. The specificity is given by a short antisense RNA sequence that hybridizes with the initial part of the protein encoding mRNA.
Why is it an important discovery?
Very little used to be known about "long, non-coding" RNAs and this new research sheds light on some of these molecules. "We focused on one gene, Uchl1, whose mutations are linked to some hereditary types of Parkinson's disease," stated Stefano Gustincich, Professor at SISSA. "We have seen that the non-coding antisense RNA matched to this gene is made up of two fragments, the real antisense fragment matching with the sense RNA that codifies the protein and the SineB2 sequence. The antisense fragment has the function of a 'lock' into which the key of the coding RNA specific for that gene is inserted, while the other one has a stimulating function on protein synthesis."
If you change the antisense fragment with the analogous of another gene, the SineB2 sequence maintains its stimulating function on the new gene. "This is important," explained Gustincich "because it means that the action of sineB2 could be used to stimulate protein production for therapeutic use – any protein – in industrial synthesis processes."
"We are delighted to see that there is one more function for long non-coding RNAs," says Piero Carninci, Team Leader at RIKEN OSC. "Since the initial discovery that the majority of the genome produces so many non-coding RNAs, there has been a general skepticism related to the possible function of these RNAs. This is a milestone study identifying a novel class of non-coding RNAs which have a key regulatory function, enhancing protein translation. Additionally, this function is mediated by repetitive elements, so far generally considered the 'junk' fraction of the genome, suggesting that the concept that most of the genome is 'junk' should be revisited. After all, there may be function embedded in any part of the genome, which we do not yet understand."
RIKEN and the RIKEN venture company TransSINE Technologies are committed to exploit commercial applications of this specific structure of the antisense RNA.
These RNAs, called SINEUPs, can be engineered to stimulate translation of other proteins, by changing the overlapping antisense region to target any protein of industrial or therapeutic interest. Initial target proteins will include therapeutic proteins, like antibodies or other soluble factors, as well as other basic studies to understand gene function by overexpression of proteins. RIKEN believes that this work can be broadly used.
More information: Claudia Carrieri, Marta Biagioli, Laura Cimatti, Anne Beugnet, Isidre Ferrer, Silvia Zucchelli, Stefano Biffo, Allistar Forrest, Piero Carninci, Elia Stupka, and Stefano Gustincich. "Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat." Nature, 2012, DOI: 10.1038/nature11508