SNPs associated with breast cancer risk alter binding affinity for pioneer factor FOXA1
Dartmouth scientists showed that more than half of all the SNPs associated with breast cancer risk are located in distant regions and bound by FOXA1, a protein required for estrogen receptor-α (ER) function according to a paper published in the journal Nature Genetics in November.
Jason Moore, PhD, a Third Century Professor of genetics, director of the Institute for Quantitative Biomedical Sciences, and associate director for bioinformatics at Dartmouth-Hitchcock Norris Cotton Cancer Center, and other researchers used a new methodology that combines cistromics, epigenomics, and genotype imputation to annotate the non-coding regions of the geneomie in breast cancer cells and systematically identify the function nature of SNPS associated with breast cancer risk.
"Understanding the biology behind the genetic risk factors opens the door to identifying new drug targets," said Dr. Moore. Results showed that, for breast cancer, the majority of risk-associated SNPs modulate FOXA1 binding. First, they are in complete linkage disequilibrium (LD) with SNPs localized to sites of FOXA1 binding, and, second, these linked SNPs are capable of changing the recruitment of FOXA1 in a significant manner.
Pioneer factors, such as FOXA1, and lineage-specific factors, such as ESR1, underlie the transcriptional programs that establish cell identity. Accordingly, researchers indicated that the majority of SNPs that can disrupt normal breast cell identity modulate the binding of the FOXA1 pioneer factor.
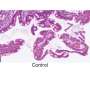
The mechanisms underlying breast cancer risk–associated SNPs are unknown. As with most other complex traits, these risk-associated SNPs map to the non-coding regions of the genome. Researchers demonstrated that breast cancer–associated SNPs are enriched for FOXA1 and ESR1 transcription factor–binding sites and H3K4me1 histone modification. Enrichment is dependent on factor, cell type and cancer type. The body of evidence supporting regulatory mechanisms for GWAS-identified risk-associated SNPs is steadily growing. Heterozygous sites with differential allelic occupancy within 100 bp of transcription start sites have been shown to have a strong association with differential gene expression and to be enriched for GWAS-identified SNPs31. Binding of the FOXA1 pioneer factor is central for chromatin opening and nucleosome positioning favorable to transcription factor recruitment. In addition, FOXA1 is central to the establishment of the transcriptional programs that respond to estrogen stimulation in ESR1-positive breast cancer cells.