Genomes of cholera bacteria from Haiti confirm epidemic originated from single source
The strain of cholera that has sickened thousands in Haiti came from a single source and was not repeatedly introduced to the island over the past three years as some have thought, according to a new study published in mBio, the online open-access journal of the American Society for Microbiology.
The results of this latest study are consistent with earlier findings that indicate Vibrio cholerae bacteria were introduced to Haiti by United nations soldiers between July and October 2010, when Nepalese soldiers arrived to assist recovery efforts after the January 2010 earthquake in that country. The genome sequences of V. cholerae strains from Haiti reveal they have not gained any new genetic material since their introduction and that they have a limited ability to acquire genes from other organisms through a process called transformation.
This new information may help public health authorities understand future cholera outbreaks in Haiti and elsewhere, according to the authors. "The use of high resolution sequence data that is amenable to evolutionary analysis will greatly enhance our ability to discern transmission pathways of virulent clones such as the one implicated in this epidemic," write the authors.
The earthquake in January 2010 killed tens of thousands of Haitians, and it was followed several months later by an outbreak of cholera, a disease that had never before been documented in Haiti. Studies of the outbreak indicate that poor sanitation at a United Nations camp resulted in sewage contamination of local water supplies, and phylogenetic analysis of the Haiti V. cholerae strains and strains from around the globe indicate the strain was most likely accidentally brought to the camp by U.N. troops from Nepal.
Earlier "fingerprinting" of Haiti's V. cholerae isolates using pulse-field gel electrophoresis (PFGE) has shown the bacterium has changed somewhat since the epidemic began in October 2010, but because of the nature of PFGE, the significance of those changes was not known. Were the changes meaningful? Were the bacteria gaining or losing genes that could impact the course of disease? Did they gain genes from other bacteria in the environment? Are their genomes rearranged? The answers could make a difference in the severity of future outbreaks.
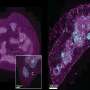
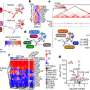
The authors of the study in mBio set out to study in greater detail how V. cholerae may have evolved since its introduction to the island nation, and whether it has acquired genes that bestow new abilities. They sequenced the genomes of 23 different V. cholerae isolates from Haiti that represent multiple PFGE "fingerprint" patterns and were taken from a variety of locations and at various time points during the epidemic.
When compared with the genome sequences of V. cholerae strains from around the world, the Haiti isolates and three Nepal isolates are tightly related, forming a monophyletic group to which no other genome sequences belong.
This result indicates that "Nepalese isolates are the closest relatives to the Haiti strain identified to date, even when placed into a phylogeny with a larger collection of isolates representing recent cholera epidemics," write the authors. This means that the outbreak originated from a single introduction of bacteria, and PFGE variants arose from gradual evolution of the organisms, not from any secondary introduction.
The Haiti strains also have a limited ability to acquire new genes through the process of transformation, by which genetic material is picked up from other bacteria or from the environment. There is some evidence that transformation is an important mechanism for bacteria to acquire the necessary abilities to adapt to a particular environment, so the fact that the Haiti strains are deficient in this respect raises the question of whether they will be able to adapt to life in Haiti or if they might go extinct once the epidemic has ended.
The Haiti isolates belong to a type of V. cholerae called "Atypical El Tor" strains, a group that, in locations in Asia and Africa, has managed to acquire multidrug resistance and enhanced virulence traits that result in higher infection rates and harsher symptoms. The authors argue that to avert larger and more difficult to treat outbreaks of cholera, it is necessary to track the ongoing and unpredictable evolution of the organism in Haiti and elsewhere with surveillance of V. cholerae via tools like whole genome sequencing.