Gene sequencing project discovers common driver of a childhood brain tumor
The St. Jude Children's Research Hospital-Washington University Pediatric Cancer Genome Project has identified the most common genetic alteration ever reported in the brain tumor ependymoma and evidence that the alteration drives tumor development. The research appears February 19 as an advanced online publication in the scientific journal Nature.
The results provide a foundation for new research to improve diagnosis and treatment of ependymoma, the third most common brain tumor in children. St. Jude has begun work to translate the discovery into new treatments for a disease that remains incurable in 40 percent of young patients. The findings should also aid efforts to understand and intervene against other cancers, including adult tumors.
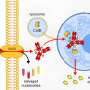
The newly discovered alteration involves a gene named RELA. The gene plays a pivotal role in the NF-kappa-B pathway, a signaling system in cells that regulates inflammation. Researchers have long recognized that this pathway is inappropriately switched on in many adult tumors. This study marks the first time scientists have found a repeated mistake—a gene alteration–in the central part of the pathway in brain cancer.
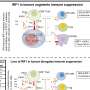
In this study, 70 percent of young patients with ependymomas in the front part of the brain carried the RELA alteration and few other genetic changes. The alteration was not found in ependymomas in other regions of the brain.
"In this study, we demonstrate for the first time that a frequent mutation in the heart of the NF-kappa-B pathway is sufficient to transform normal brain cells into cancer cells and drive tumor development," said co-corresponding author Richard Gilbertson, M.D., Ph.D., director of the St. Jude Comprehensive Cancer Center. "This should help us to understand how abnormal NF-kappa-B activity drives cancer and to develop new treatments to block that activity."
The alteration fuses RELA with parts of another gene, C11orf95, in a process called translocation. The translocation produces abnormal proteins that rapidly cause fatal brain tumors in mice. These tumors resembled the human disease.
"This is an exciting finding, not only for understanding the biology of a rare and particularly devastating childhood brain cancer, but also for understanding how it might be effectively treated," said co-author Richard K. Wilson, Ph.D., director of The Genome Institute at Washington University School of Medicine in St. Louis.
St. Jude is now leading an international study to determine if the C11orf95-RELA translocation might help predict the outcome for ependymoma patients. St. Jude has also developed a test to identify tumors that carry the translocation. Co-corresponding author David Ellison, M.D., Ph.D., St. Jude Department of Pathology chair, led that effort.
An analytic tool called CICERO developed by St. Jude researchers played a key role in identifying the translocation. Co-corresponding author Jinghui Zhang, Ph.D., an associate member of the St. Jude Department of Computational Biology, led the team that developed CICERO.
Finding the translocation required sifting through 246 billion pieces of genetic information that contain the complete genetic code of the tumor as well as the normal DNA from 41 young patients with ependymoma. The researchers also studied the RNA in 77 ependymomas. The DNA in cells contains the code for each gene, while RNA transmits this information to the part of the cell that makes proteins. By sequencing both the DNA and RNA, scientists can see which genes are turned on and are making abnormal products such as the RELA translocation.
Using CICERO, researchers found abnormalities in RNA that led them to the C11orf95-RELA translocation. The fusion gene was created when a piece of chromosome 11 that houses both the C11orf95 and RELA genes was shattered and incorrectly reassembled.
The result is one of the most commonly occurring translocations ever reported in brain tumors. Of the 41 ependymomas in this study that began in the front part of the brain, 29 tumors had the translocation and made RELA fusion proteins. "The fact the alteration results in abnormal proteins offers a potential new therapeutic target, which is significant for ependymoma," Gilbertson said.
Researchers are working to understand how the fusion proteins cause cancer. Evidence suggests that C11orf95 plays a key role by altering the way that RELA moves through the cell and performs its normal functions. Investigators also discovered translocations involving other genes that appear to drive ependymoma.
The study was part of the Pediatric Cancer Genome Project, which has sequenced the complete normal and tumor genomes of 700 young cancer patients. The project was launched in 2010 to harness advances in genome sequencing technology to improve understanding and treatment of some of the most aggressive and least understood childhood cancers.
More information: C11orf95–RELA fusions drive oncogenic NF-kappa-B signalling in ependymoma, DOI: 10.1038/nature13109