Tumor-suppressor connects with histone protein to hinder gene expression
A tumor-suppressing protein acts as a dimmer switch to dial down gene expression. It does this by reading a chemical message attached to another protein that's tightly intertwined with DNA, a team led by scientists at The University of Texas MD Anderson Cancer Center reported at the AACR Annual Meeting 2014.
The findings, also published in the journal Nature on April 10, provide evidence in support of the "histone code" hypothesis. The theory holds that histone proteins, which combine with DNA to form chromosomes, are more intimately involved in gene expression than their general role of facilitating or hindering gene activation suggests.
The researchers found that high expression of the tumor-suppressor ZMYND11 is associated with longer survival for patients with triple-negative breast cancer.
"This study, for the first time, identifies a novel role of a histone variant protein in regulating gene transcription aside from its established roles," said senior author Xiaobing Shi, Ph.D., assistant professor of Biochemistry and Molecular Biology at The University of Texas MD Anderson Cancer Center.
"We also found that this variant, H3.3, is modified by methylation to create a specific epigenetic landscape that is accommodated by the tumor-suppressing protein ZMYND11. The protein in turn blocks gene activation," Shi said. "This is exactly the type of combined effect predicted by the histone code hypothesis."
Methylation, the attachment of a methyl group to a gene or protein, and other types of histone modifications are considered epigenetic factors, which modify a gene's behavior without changing its DNA coding.
Shi and colleagues found that the protein ZMYND11 "reads" the modified histone H3.3 by connecting to it where a tri-methyl chemical group binds to H3.3. From this position, Shi said, ZMYND11 thwarts a step in gene activation called elongation, inhibiting cancer growth.
ZMYND11 expression shrinks tumors in mice
Extensive structural analysis established that the ZMYND11- methylated H3.3 combination hunkers down in the gene's DNA.
"We knew ZMYND11 was a candidate tumor-suppressor because it's down-regulated in a number of human cancers, including breast cancer," Shi said.
Overexpression of ZMYND11 in an osteosarcoma cell line and a triple-negative breast cancer cell line inhibited tumor growth. Versions of ZMYND11 that could not bind to the trimethyl group on H3.3 did not suppress cancer cell growth or survival.
In a mouse model of triple-negative breast cancer, mice injected with cancer cells that over-express ZMYND11 had tumor volumes of less than 50 cubic millimeters while control mice and those injected with cells expressing ZMYND11 deficient for binding to the methyl group had tumor volumes ranging from 150 to 400 cubic millimeters at eight weeks.
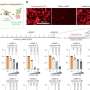
When the researchers knocked the ZMYND11 gene down in an osteosarcoma cell line, they found 268 genes had increased expression while 370 genes were down-regulated. Further analysis showed some of the activated genes were enriched in small cell lung cancer and other cancer-promoting pathways.
This pointed to a role in both the repression and activation of gene transcription.
Associated with longer survival for triple-negative breast cancer patients
Gene activation begins when a transcription factor connects with the gene's promoter region. An enzyme called polymerase II then moves along the gene's DNA like a zipper, reading the DNA to produce a strand of RNA, a process called elongation. This ends when the polymerase hits the gene's stop signal.
Since their structural research had shown the ZMYND11/H3.3 combination localized in a gene's DNA rather than its promoter region, the team hypothesized that it fine-tunes gene expression during elongation rather than acting as an on-off switch in the gene's promoter region.
Subsequent experiments showed that the polymerase was more active in the gene body when ZMYND11 was suppressed, particularly on genes that ZMYND11 inhibited.
An analysis of ZMYND11 levels in the tumors of 120 triple-negative breast cancer patients showed that those with high levels of the protein had an 80 percent probability of surviving for 10 years while those with low levels had a 50 percent probability.
The researchers are interested in further elucidating the detailed mechanisms by which ZMYND11 controls transcription elongation.
"Although we know that ZMYND11 controls RNA polymerase II travel ratio in the gene body, we still don't know how this protein, which does not physically interact with polymerase II, actually achieves this regulation," Shi said.
"The next thing to do is to generate a knockout mouse model for further in vivo analysis, as the ultimate goal of our research is to move from bench to bedside, and generating a mouse model is a key step during this long journey," Shi said.