Predicting your risk of infection
Research publishing this week in PLOS Computational Biology analyses the livestock trade in Italy and sexual encounters in a Brazilian prostitution service to find a correlation between loyalty and infection risk.
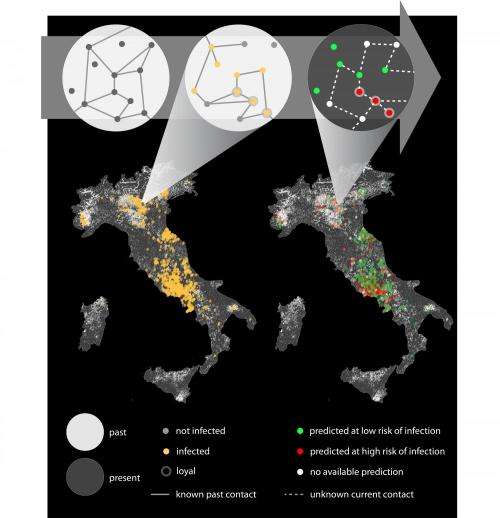
The French team at Inserm, in collaboration with ISI Foundation and Istituto Zooprofilattico Abruzzo-Molise in Italy, has shown that it is possible to use past contact data to predict the risk of infection in emerging outbreaks.
The study analyzes contact data between animal premises in Italy following livestock trade movements, and time-stamped sexual encounters in a high-end prostitution service in Brazil. The first is relevant for the spread of livestock diseases, and the second underlies the potential spread of sexually transmitted diseases.
By introducing a measure of loyalty in the way farmers or prostitution clients tend to establish their contacts with the same individuals over time (for livestock trade or sexual behavior), the authors uncovered a significant correlation between the loyalty of an individual and his/her risk of infection.
Contact patterns for disease transmission contribute to a detailed understanding of the disease spread and risk of infection, however they are often not readily available for analysis due to their time-evolving nature.
In a transmissible disease epidemic, these findings can be used to guide the prioritization of available interventional resources to efficiently control the disease spread. The general nature of the study makes its results applicable to a wide spectrum of possible epidemic scenarios.
More information: Valdano E, Poletto C, Giovannini A, Palma D, Savini L, Colizza V (2015) Predicting Epidemic Risk from Past Temporal Contact Data. PLoS Comput Biol 11(3): e1004152. DOI: 10.1371/journal.pcbi.1004152