Researchers identify molecular mechanism responsible for making malaria parasites drug-resistant
University of Notre Dame researchers led an international team to identify a molecular mechanism responsible for making malaria parasites resistant to artemisinins, the leading class of antimalarial drugs.
According to the World Health Organization's 2014 World Malaria Report, there are an estimated 198 million cases of malaria worldwide with 3.3 billion people at risk for contracting the infection. Although the impact of malaria is still significant, the statistics reflect a considerable reduction in the global malaria burden. Since 2010, disease transmission has been reduced by 30 percent and mortality due to malaria has decreased by almost half.
Artemisinins are powerful drugs that have the most rapid action of all current drugs against Plasmodium falciparum, the parasite species that causes the most dangerous form of malaria. Artemisinin combination therapies (ACTs) are now standard treatment worldwide for P. falciparum malaria. Unfortunately, resistance to artemisinin has been detected in five countries across Southeast Asia. Along the Cambodia-Thailand border, P. falciparum is now resistant to most available antimalarial drugs. Artemisinin resistance poses a serious global threat to malaria control and elimination.
"There are two phases of blood stage malaria infection. In the first phase, the 'ring' parasite stage circulates in the bloodstream, and in the second phase, the 'mature' parasite stage sequesters in the tissues of the body," explained Kasturi Haldar, the Rev. Julius A. Nieuwland Professor of Biological Sciences and the James C. Parsons and Carrie Ann Quinn Director of the Boler-Parseghian Center for Rare and Neglected Diseases. "Artemisinins are highly effective in treating malaria quickly because they target the first ring stage. When patients take the medication, their fevers reduce quickly, and the parasite is eliminated rapidly."
Although artemisinins have been widely used and investigated, no one fully understood how they worked or why clinical resistance has emerged. Laboratory studies showed that artemisinins were always active against the mature parasite stages that sequester in tissues, but clinicians were observing resistant infections from patients who were still in early stage of infections. In addition, previous genome wide association studies of P. falciparum identified genes associated with artemisinin resistance, but it was unknown how the genes worked. This Notre Dame-led study identified both the target of artemisinins in the clinically affected ring stages and how a gene named PfKelch13, a dominant marker used to track the parasite's resistance, causes artemisinin resistance.
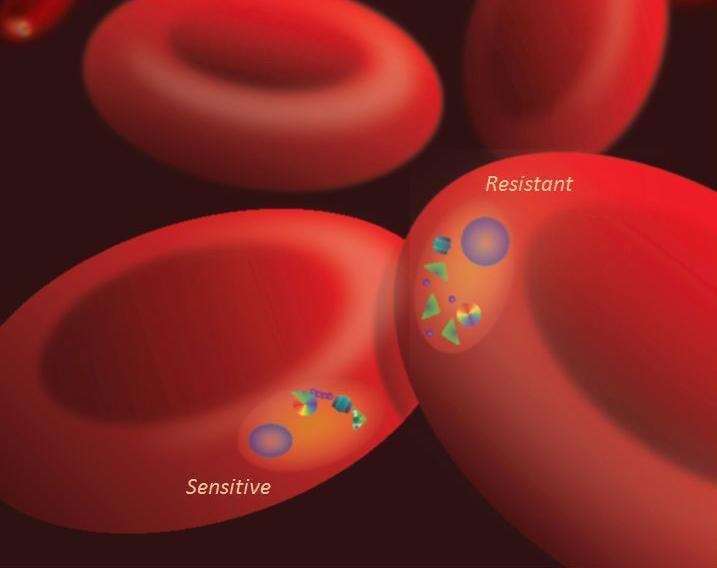
"We observed that levels of a lipid called phosphatidylinositol-3-phosphate (PI3P) were higher in artemisinin-resistant P. falciparum than artemisinin-sensitive strains," said Alassane Mbengue, a postdoctoral associate in biological sciences and co-first author on the study. "This lipid is produced by an enzyme called PfPI3K. We found that artemisinins block this kinase from producing PI3P lipids."
"We also discovered that the amount of the kinase present in the parasite is controlled by the gene PfKelch13," Mbengue said. "Mutation in the gene increases the kinase levels, which in turn increases PI3P lipid levels. The higher the level of PI3P lipids present in the parasite, the greater the level of artemisinin resistance. We also studied the lipid levels in parasites without the gene mutation and observed that when PI3P lipid levels were increased artificially, the parasites still became proportionately resistant."
"Our results are especially significant because we studied clinical parasites from Cambodia, where artemisinin resistance is highly prevalent," explained Souvik Bhattacharjee, research assistant professor of biological sciences and co-first author of the paper. "We collaborated with researchers and clinicians in Asia, Europe and the U.S. The local collaborators at the Boler-Parseghian Center for Rare and Neglected Diseases include Rob Stahelin at IUSM, Olaf Wiest in the Department of Chemistry and Biochemistry, the Computer Assisted Molecular Design Core, Shahir Rizk in the Department of Biological Sciences and the Genomics and Bioinformatics Core. This was true cooperation over several years at both the international and the local levels."
When asked about the next steps for this research, Bhattacharjee said, "There are presently two options for overcoming artemisinin resistance. Working with our colleagues at Notre Dame, Eli Lilly & Co. and the Medicines for Malaria Venture, we need to find drugs that kill the parasite by blocking the function of the kinase from making the PI3P lipid or disrupting the production of the kinase itself."
More information: "A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria" Nature (2015) DOI: 10.1038/nature14412