Scientists find minor flu strains pack bigger punch
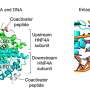
Minor variants of flu strains, which are not typically targeted in vaccines, carry a bigger viral punch than previously realized, a team of scientists has found. Its research, which examined samples from the 2009 flu pandemic in Hong Kong, shows that these minor strains are transmitted along with the major strains and can replicate and elude immunizations.
"A flu virus infection is not a homogeneous mix of viruses, but, rather, a mix of strains that gets transmitted as a swarm in the population," explains Elodie Ghedin, a professor in New York University's Department of Biology and College of Global Public Health. "Current vaccines target the dominant strains, because they are the ones that seem to infect the largest number of individuals. But our findings reveal an ability of minor strains to elude these vaccines and spread the virus in ways not previously known."
The other co-authors of the paper, which appears in the journal Nature Genetics, included: Leo Poon, an associate professor at the University of Hong Kong; Tim Song, a graduate student at the University of Pittsburgh School of Medicine at the time of the study and now a computational biologist at NYU; and researchers at the J. Craig Venter Institute, the University of Sydney, and the Icahn School of Medicine at Mt. Sinai.
It's long been known that the Influenza A virus is marked by a high level of genetic diversity. However, our knowledge largely stems from the dominant strain, which vaccines aim to combat. Less understood is the diversity of the minor strains and how it passes between individuals—thus revealing these strains' ability to spread the virus.
The scientists aimed to determine how many viral particles are transmitted when afflicted with the flu as well as the number of them able to replicate when they transmit.
To examine this phenomenon, Ghedin, part of NYU's Center for Genomics & Systems Biology, and her colleagues performed whole genome deep sequencing of upper nasal cavity swabs taken from confirmed 2009 Hong Kong flu cases and from their household contacts.
Using sophisticated sequencing methods, the team could not only identify variants in flu strains, but also quantify what was being transmitted between infected individuals.
Their results showed that, as expected, most carried the dominant virus—H1N1 or H3N2. But, in addition, all carried minor strains and variants of the major and minor strains. What was surprising was how readily these variants were transmitted across the studied individuals.
"The combination of unique data, sequencing approaches and mathematical methods create a nuanced picture of the transmission of diversity during a pandemic," notes study co-author Benjamin Greenbaum, an assistant professor at the Tisch Cancer Institute at the Icahn School of Medicine at Mount Sinai.
"We were able to look at the variants and could link individuals based on these variants," adds Ghedin. "What stood out was also how these mixes of major and minor strains were being transmitted across the population during the 2009 pandemic—to the point where minor strains became dominant."
More information: Quantifying influenza virus diversity and transmission in humans, DOI: 10.1038/ng.3479